January 03, 2018 by Kelly Hondula
Photos, as a source of data, or to aid in the interpretation of data, can be a useful addition to RShiny applications. Here are two examples of using photo data: one that displays images from URLs, and another that uses species names to find pictures of animals.
The first situation was using a social media dataset (images from Flickr) that was a table where each row contained information about a photograph, and columns contained data about that photo. One column contained URLs to the image file. The goal was to be able to browse through each image and potentially update other information in the table pertaining to that image. This can be done in Shiny with an interactive DataTable, where clicking on a row displays an image and allows for making changes to its associated data.
When a row in the table is clicked, the ID of that row becomes an input variable that can be used in the shiny server. That variable can then be used to select the URL text string in the “image_url” column of the image_data dataframe. That URL is then enclosed within html img tags to create as a text object called “image1”.
output$image1 <- renderText({
c('<img src="', image_data[input$image_data_rows_selected, "image_url"], '">')
})
Output objects rendered within the shiny server need to be paired with a corresponding output function in the user interface. Since the text output object “image1” is actually an html string, using it with htmlOutput will interpret the html and display the image wherever it is placed in the app!
out1 <- htmlOutput("image1")
See a minimal example on github here.
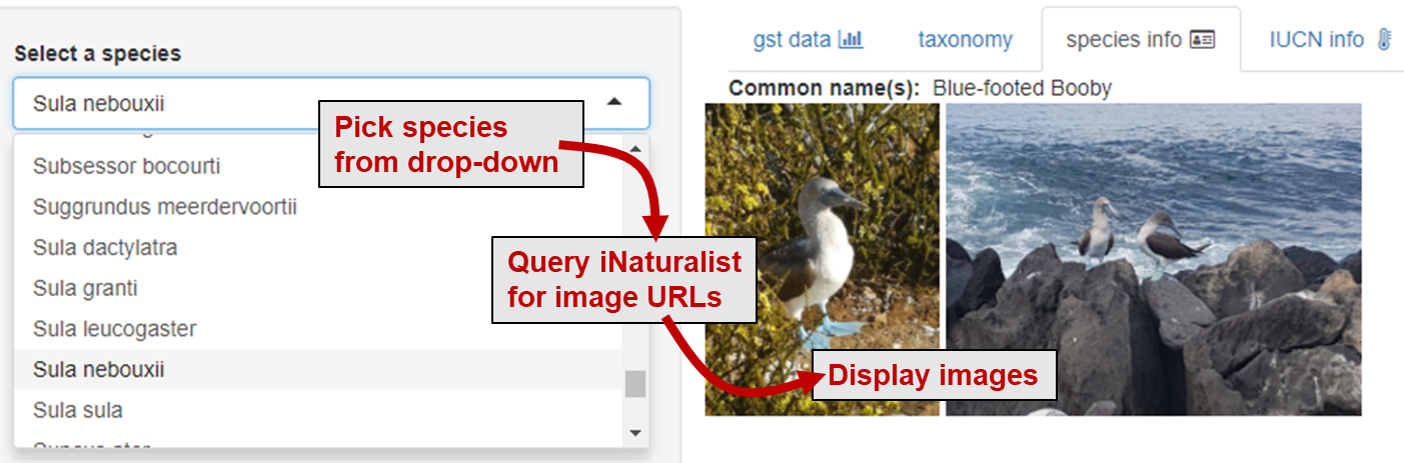
Another shiny app that used images to aid in data exploration shiny app was based on a large biodiversity data set. The main purpose of the app was to display country-level time series data for a selected species—but with thousands of different taxa, exploring the dataset based on latin names proved difficult! So, I wanted the app to display some contextual information about the selected species aside from its latin name and the time series data. This is possible with functions from the rinat package, whichs draws on iNaturalist data—which includes urls of user-uploaded images based on taxa names.
inat <- reactive({
get_inat_obs(taxon_name = input$selected_species)
})
The function get_inat_obs() retrieves a dataframe of species observations based on the species chosen in a drop-down menu. One of the columns contains image URLs, which can then be used to create an output image object.
output$image1 = renderUI({
tags$img(src=inat()$image_url[1], height = "200px")
})
The taxize and wikitaxa packages also have useful functions for determining the common name, taxonomy, conservation status, geographic distribution, and more.