Documenting and Publishing your Data
Lesson 10 with Rachael Blake
Lesson Objectives
- Review key concepts of making your data tidy
- Discover the importance of metadata
- Learn what a data package is
- Prepare to publish your data from R
Specific Achievements
- Create metadata locally
- Build data package locally
- Learn how data versioning can help you collaborate more efficiently
- Practice uploading data package to repository (exercise outside lesson)
What is a data package?
A data package is a collection of files that describe your data.
There are two essential parts to a data package: data and metadata.
Data packages also frequently include 1) code scripts that clean, process, or perform statistical anslyses on your data, and 2) visualizations that are direct products of coded anlyses. The relationships between these files are described in the provenance information for a data package. We will get into more details on these components in this lesson.
Why package and publish your data?
-
Data is a valuable asset - easily publish your data now or in the future
-
Efficient collaboration - improve sharing data with collaborators now
-
Credit for your work
-
Funder requirement
-
Publisher requirement
-
Open, Reproducible Science
Integrating data documentation
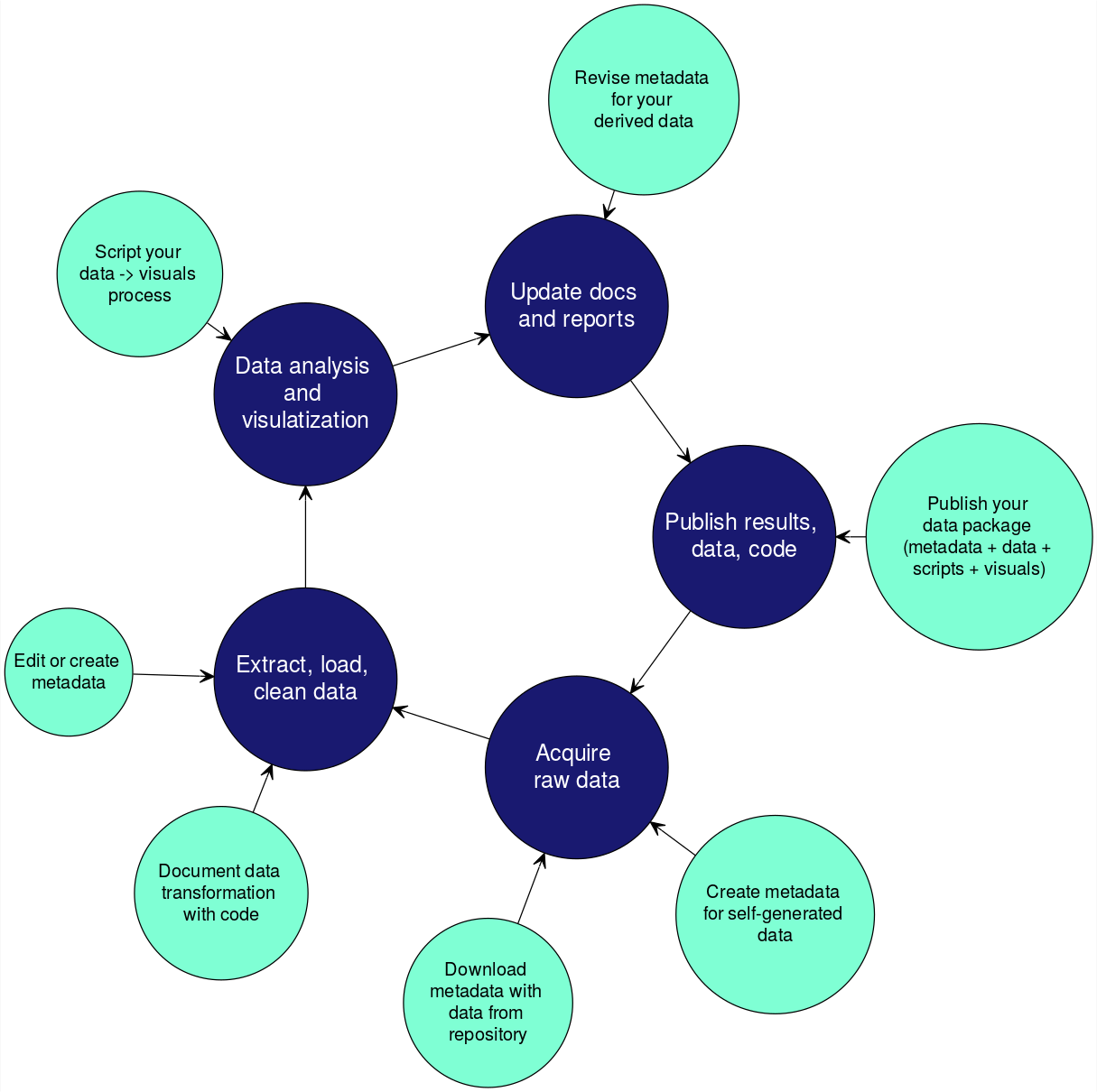
It is important to incorporate documentation of your data throughout your reproducible research pipeline.
Preparing Data for Publication
Both raw data and derived data (data you’ve assembled or processed in some way) from a project need to be neat and tidy before publication. Following principles of tidy data from the beginning will make your workflows more efficient during a project, and will make it easier for others to undertand and use your data once published. However, if your data isn’t yet tidy, you can make it tidier before you publish it.
Consistency is key when managing your data!
-
At a minimum, use the same terms to refer to the same things throughout your data.
- “origin”, not “starting place”, “beginning location”, and “source”
- “Orcinus orca”, not “killer Whale”, “Killer whale”, “killer.whale”, and “orca”
-
Consider using a discipline-wide published vocabulary if appropriate (ex: hydrological controlled vocab.)
Naming data files
Bad: “CeNsus data*2ndTry 2/15/2017.csv”
Good: “census_data.csv”
library(tidyverse)
stm_dat <- read_csv("data/StormEvents.csv")
Formatting data
- One table/file for each type of observation.
- Each variable has its own column.
- Each observation has its own row.
- Each value has its own cell.
Downstream operations/analysis require tidy data.
These principles closely map to best practices for “normalization” in database design. R developer Hadley Wickham further describes the priciples of tidy data in this paper (Wickham 2014).
You can work towards ready-to-analyze data incrementally, documenting the intermediate data and cleaning steps you took in your scripts. This can be a powerful accelerator for your analysis both now and in the future.
Let’s do a few checks to see if our data is tidy.
- make sure data meet criteria above
- make sure blanks are NA or some other standard
> head(stm_dat)
> tail(stm_dat)
- check date format
> str(stm_dat)
- check case, white space, etc.
> unique(stm_dat$EVENT_NARRATIVE)
Outputting derived data
Because we followed the principle of one table/file for each type of observation, we have one table to be written out to a file now.
dir.create('storm_project', showWarnings = FALSE)
write_csv(stm_dat, "storm_project/StormEvents_d2006.csv")
Versioning your derived data
Issue: Making sure you and your collaborators are using the same version of a dataset
One solution: version your data in a repository (more on this later)
What is Metadata?
Metadata is the information needed for someone else to understand and use your data. It is the Who, What, When, Where, Why, and How about your data.
This includes package-level and file-level metadata.
Common components of metadata
Package-level:
- Data creator
- Geographic and temporal extents of data (generally)
- Funding source
- Licensing information (Creative Commons, etc.)
- Publication date
File-level:
- Define variable names
- Describe variables (units, etc.)
- Define allowed values for a variable
- Describe file formats
Metadata Standards
The goal is to have a machine and human readable description of your data.
Metadata standards create a structural expression of metadata necessary to document a data set.
This can mean using a controlled set of descriptors for your data specified by the standard.
Without metadata standards, your digital data may be irretrievable, unidentifiable or unusable. Metadata standards contain definitions of the data elements and standardised ways of representing them in digital formats such as databases and XML (eXtensible Markup Language).
These standards ensure consistent structure that facilitates data sharing and searching, record provenance and technical processes, and manage access permissions. Recording metadata in digital formats such as XML ensures effective machine searches through consistent sturctured data entry, and thesauri using controlled vocabularies.
Metadata standards are often developed by user communities.
For example, EML was developed by ecologists to describe environmental and ecological data.
Therefore, the standards can vary between disciplines and types of data.

Some examples include:
- General: Dublin Core
- Ecological/Environmental/Biological: EML, Darwin Core
- Social science: DDI, EAD
- Geospatial/Meterological/Oceanographic: ISO 19115, FGDC/CSDGM (no longer current)
Creating metadata
Employer-specific mandated methods (ex: USGS)
Repository-specific methods
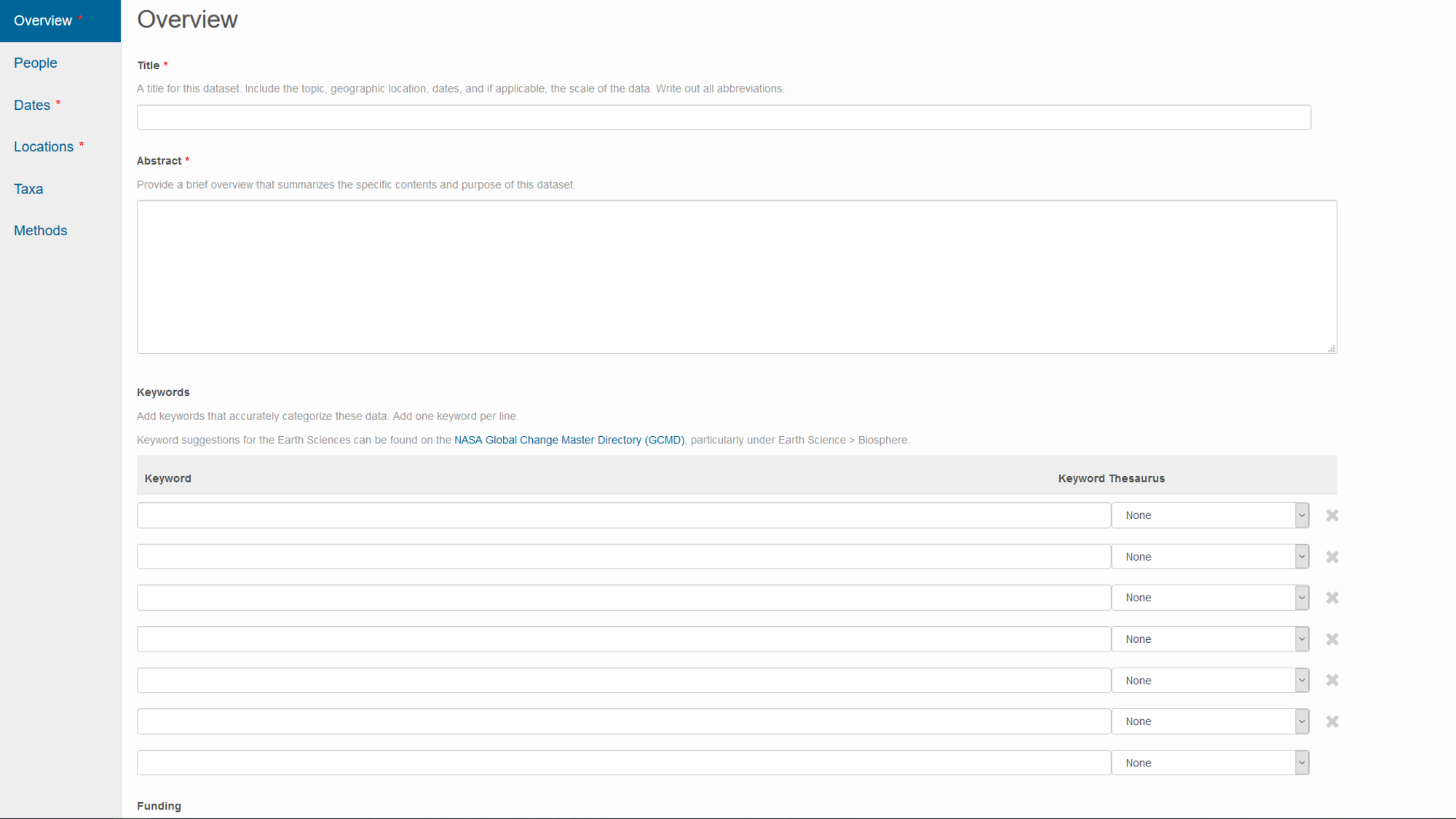
Repository websites frequently guide you through metadata creation during the process of uploading your data.
- website for metadata entry on the Knowledge Network for Biocomplexity repository
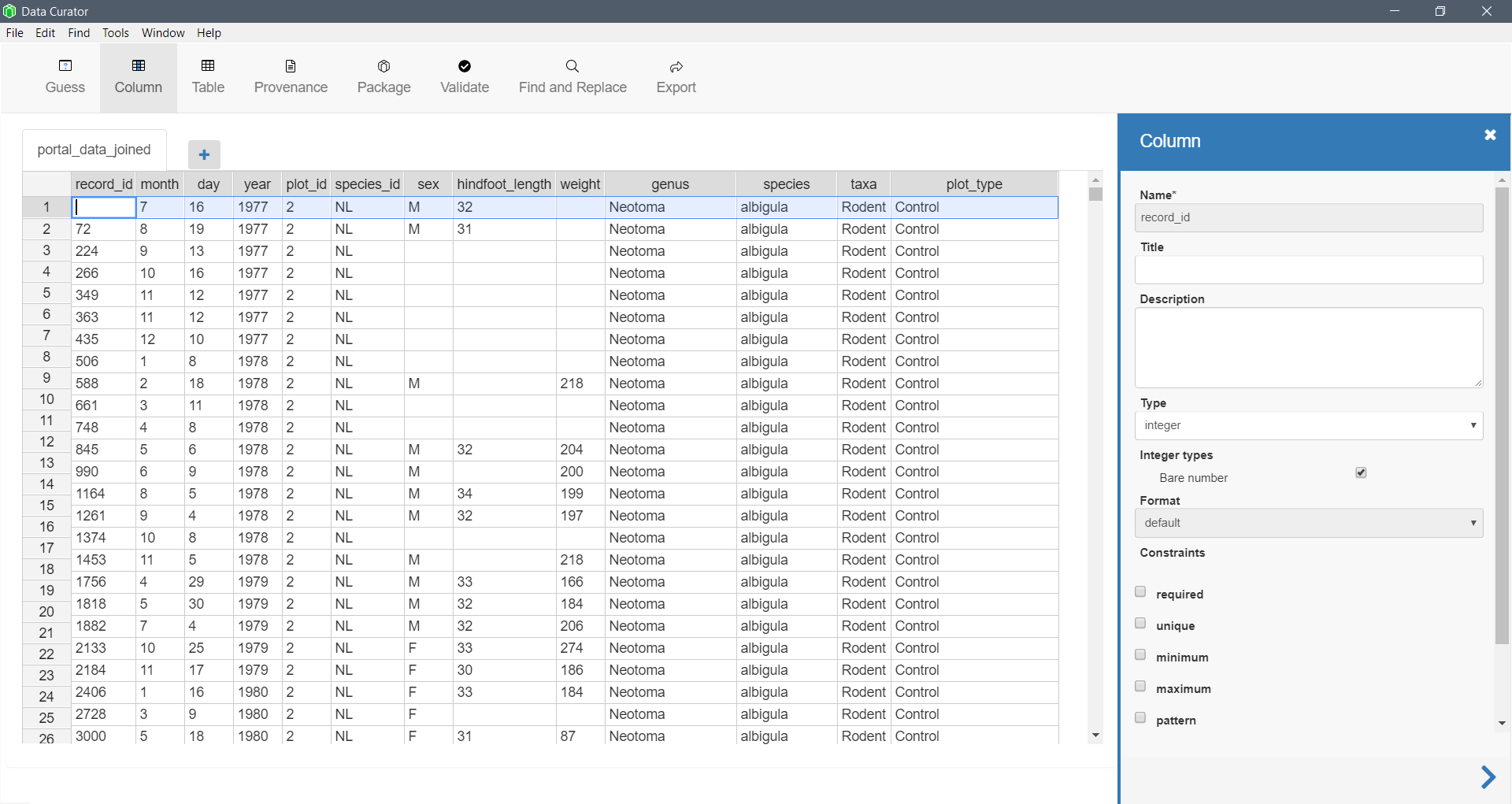
Stand-alone software
Software designed for data curation often has more features and options. This software from the Queensland Cyber Infratstructure Foundation supports metadata creation, provenance, and data packaging.
Coding
Many methods for documenting and packaging data are also available in R packages developed by repository managers. For example, EMLassemblyline was developed by the Environmental Data Initiative to facilitate documenting data in EML, and preparing data for publication.
- R packages (EML, dataspice, emld), EMLassemblyline
Example of coding up some metadata
| R Package | What does it do? |
|---|---|
dataspice |
creates metadata files in json-ld format |
emld |
aids conversion of metadata files between EML and json-ld |
EML |
creates EML metadata files |
jsonlite |
reads json and json-ld file formats in R |
We’ll use the dataspice package to create metadata in the EML metadata standard.
library(dataspice)
Create data package templates.
create_spice(dir = "storm_project")
Look at your storm_project folder, and see the CSV template files.
Describe the package-level metadata.
The templates are empty, so now we need to populate them. Add extent, coverage, license, publication, funder, keywords, etc.
We can get the temporal and geographic extent information using the range() function.
> range(stm_dat$YEAR)
>
> range(stm_dat$BEGIN_LAT, na.rm=TRUE)
> range(stm_dat$BEGIN_LON, na.rm=TRUE)
This extent information can now be added to the biblio.csv metadata file.
This function opens a Shiny app interface where you can enter and save information in the CSV templates. You could also edit these CSV files in another software program.
edit_biblio(metadata_dir = "storm_project/metadata")
You may need to click the stop sign in your console to stop the edit Shiny app interface.
Describe the creators of the data.
edit_creators(metadata_dir = "storm_project/metadata")
You may need to click the stop sign in your console to stop the edit Shiny app interface.
The prep_access() function tries to discover the metadata for itself.
prep_access(data_path = "storm_project",
access_path = "storm_project/metadata/access.csv")
The edit_access() function can be used to edit the generated metadata, or used by itself to manually enter access metadata. Supply the file name and other access information about the data.
edit_access(metadata_dir = "storm_project/metadata")
Describe the file-level metadata.
The prep_attributes() function tries also to discover the metadata for itself.
prep_attributes(data_path = "storm_project",
attributes_path = "storm_project/metadata/attributes.csv")
The edit_attributes() function can be used to further edit the file attribute metadata.
edit_attributes(metadata_dir = "storm_project/metadata")
Write metadata file
Now we can write our metadata to a json-ld file.
write_spice(path = "storm_project/metadata")
You can also create a static website for your dataset.
build_site(path = "storm_project/metadata/dataspice.json")
Your site will build to the location “docs/index.html”.
Many repositories use EML (Ecological Metadata Language) formatting for metadata.
Now convert the json-ld file into EML format.
library(emld)
library(EML)
library(jsonlite)
json <- read_json("storm_project/metadata/dataspice.json")
eml <- as_emld(json)
write_eml(eml, "storm_project/metadata/dataspice.xml")
Publishing your data
Choosing to publish your data in a long-term repository can:
- enable versioning of your data
- fulfill a journal requirement to publish data
- facilitate citing your dataset by assigning a permanent uniquely identifiable code (DOI)
- improve discovery of and access to your dataset for others
- enable re-use and greater visibility of your work
Why can’t I just put my data on Dropbox, Google Drive, my website, etc?

Key reasons to use a repository:
- preservation (permanence)
- stability (replication / backup)
- access
- standards
When to publish your data?
Near the beginning? At the very end?
A couple issues to think about:
1) How do you know you’re using the same version of a file as your collaborator?
- Publish your data privately and use the unique ids of datasets to manage versions.
2) How do you control access to your data until you’re ready to go public with it?
- Embargoing - controlling access for a certain period of time and then making your data public.
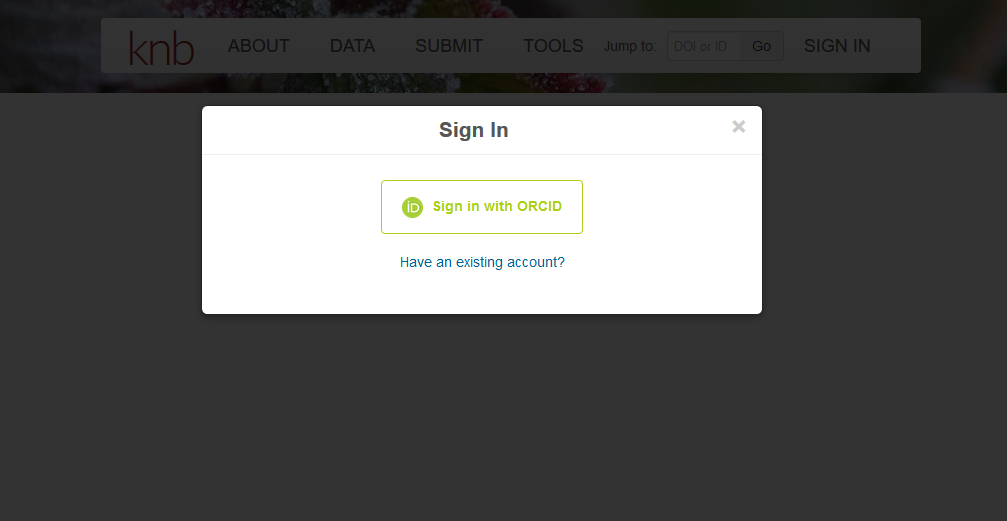
Get an ORCiD
ORCiDs identify you and link you to your publications and research products. They are used by journals, repositories, etc. and often as a log in.
To obtain an ORCiD, register at https://orcid.org.
Creating a data package
Data packages can include metadata, data, and script files, as well as descriptions of the relationships between those files.
Currently there are a few ways to make a data package:
- Frictionless Data uses json-ld format, and has the R package
datapackage.rwhich creates metadata files using schema.org specifications and creates a data package.
- DataONE frequently uses EML format for metadata, and has related R packages
datapackanddataonethat create data packages and upload data packages to a repository.
We’ll follow the DataONE way of creating a data package in this lesson.
| R Package | What does it do? |
|---|---|
datapack |
creates data package including file relationships |
uuid |
creates a unique identifier for your metadata |
We’ll create a local data package using datapack:
library(datapack)
library(uuid)
dp <- new("DataPackage") # create empty data package
Add the metadata file we created earlier to the blank data package.
emlFile <- "storm_project/metadata/dataspice.xml"
emlId <- paste("urn:uuid:", UUIDgenerate(), sep = "")
mdObj <- new("DataObject", id = emlId, format = "eml://ecoinformatics.org/eml-2.1.1", file = emlFile)
dp <- addMember(dp, mdObj) # add metadata file to data package
Add the data file we saved earlier to the data package.
datafile <- "storm_project/StormEvents_d2006.csv"
dataId <- paste("urn:uuid:", UUIDgenerate(), sep = "")
dataObj <- new("DataObject", id = dataId, format = "text/csv", filename = datafile)
dp <- addMember(dp, dataObj) # add data file to data package
Define the relationship between the data and metadata.
dp <- insertRelationship(dp, subjectID = emlId, objectIDs = dataId)
You can also add scripts and derived data files to the data package.
Create a Resource Description Framework (RDF) of the relationships between data and metadata.
serializationId <- paste("resourceMap", UUIDgenerate(), sep = "")
filePath <- file.path(sprintf("%s/%s.rdf", tempdir(), serializationId))
status <- serializePackage(dp, filePath, id=serializationId, resolveURI = "")
Save the data package to a file, using the BagIt packaging format.
Right now this creates a zipped file in the tmp directory. We’ll have to move the file out of the temp directory after it is created. Hopefully this will be changed soon!
dp_bagit <- serializeToBagIt(dp)
file.copy(dp_bagit, "storm_project/Storm_dp.zip")
Picking a repository
There are many repositories out there, and it can seem overwhelming picking a suitable one for your data.
Repositories can be subject or domain specific. re3data lists repositories by subject and can help you pick an appropriate repository for your data.
DataONE is a federation of repositories housing many different types of data. Some of these repositories include Knowledge Network for Biocomplexity (KNB), Environmental Data Initiative (EDI), Dryad, USGS Science Data Catalog
For qualitative data, there are a few dedicated repositories: QDR, Data-PASS
Though a bit different, Zenodo facilitates publishing (with a DOI) and archiving all research outputs from all research fields.
This can be used to publish releases of your code that lives in a GitHub repository. However, since GitHub is not designed for data storage and is not a persistent repository, this is not a recommended way to store or publish data.
Uploading to a repository
Uploading requirements can vary by repository and type of data. The minimum you usually need is basic metadata, and a data file.
If you have a small number of files, using a repository GUI will usually be simpler. For large numbers of files, automating uploads from R will save time. And it’s reproducible!
Data Upload Example using DataONE:
If you choose to upload your data package to a repository in the DataONE federation, the following example might be useful.
You’ll need to do two basic steps:
1) Get authentication token for DataONE (follow steps here)
2) Upload your data package using R (from vignette for dataone)
| R Package | What does it do? |
|---|---|
dataone |
uploads data package to repository |
Tokens:
Different environments in DataONE take different authentication tokens. See below for description of environments.
To get a token for the staging envrionment go here https://dev.nceas.ucsb.edu.
To get a token for production environment go here https://search.dataone.org.
Then do the following:
- Click Sign in, or Sign up if necessary
- Once signed in, move the cursor over the user name and select ‘My profile’ in the drop down menu.
- Click on the “Settings” tab.
- Click on “Authentication Token” in the menu below “Welcome”
- Click on the “Token for DataONE R” tab.
- Click “Renew authentication token” if the token you have been using has expired.
- Click on the “Copy” button below the text window to copy the authentication string to the paste buffer.
- Note the identity string and expiration date of the token.
- In the R console, paste the token string.
Set your token now
Paste your token in R console, as in the above instructions.
Set access rules
> library(dataone)
> library(curl)
> library(redland)
>
> dpAccessRules <- data.frame(subject="http://orcid.org/0000-0003-0847-9100",
+ permission="changePermission")
This gives this particular orcid (person) permission to read, write, and change permissions for others for this package
> dpAccessRules2 <- data.frame(subject = c("http://orcid.org/0000-0003-0847-9100",
+ "http://orcid.org/0000-0000-0000-0001"),
+ permission = c("changePermission", "read")
+ )
NOTE: When you upload the package, you also need to set public = FALSE if you don’t want your package public yet.
Upload data package
The first argument here is the environment - “PROD” is production where you publish your data. “STAGING” can be used if you’re not yet sure you have everything in order and want to test uploading your data package. NOTE: “PROD” and “STAGING” require different tokens. See above steps to get the correct token.
The second argument is the repository specification. If you don’t know your repository specs, look it up in this table of member node IDs: (data/Nodes.csv)
> read.csv("data/Nodes.csv")
First set the environment and repository you’ll upload to:
> d1c <- D1Client("STAGING2", "urn:node:mnTestKNB")
Now do the actual uploading of your data package:
> packageId <- uploadDataPackage(d1c, dp, public = TRUE, accessRules = dpAccessRules,
+ quiet = FALSE)
Citation
Getting a Digital Object Identifier (DOI) for your data package can make it easier for others to find and cite your data.
You can assign a DOI to the metadata file for your data package using:
First specify the environment and repository.
> cn <- CNode("PROD")
> mn <- getMNode(cn, "urn:node:DRYAD")
>
> doi <- generateIdentifier(mn, "DOI")
Now overwrite the previous metadata file with the new DOI identified metadata file
> mdObj <- new("DataObject", id = doi, format = "eml://ecoinformatics.org/eml-2.1.1",
+ file = emlFile)
Versioning Data
How do you know you’re using the same version of a data file as your collaborator?
- Do not edit raw data files! Make edits/changes via your code only, and then output a derived “clean” data file.
Potential ways forward:
-
Upload successive versions of the data to a repo with versioning, but keep it private until you’ve reached the final version or the end of the project. (see below)
-
Google Sheets has some support for viewing the edit history of cells, added or deleted columns and rows, changed formatting, etc.
Updating your data package on the repository
To take advantage of the versioning of data built into many repositories, you can update the data package (replace files with new versions).
Please see the vignettes for dataone , section on “Replace an object with a newer version”.
Provenance
Prov is tracking the inputs and outputs generated by the research process, from raw data to publication(s).
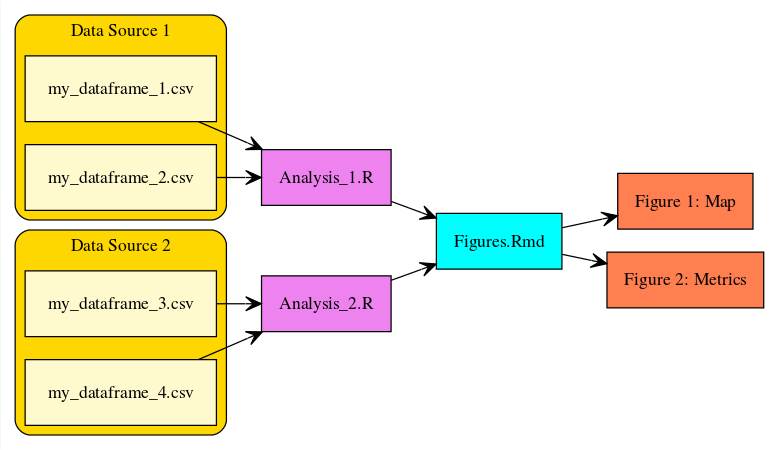
It is often very helpful to create a diagram that shows the relationships between files.
A couple examples include:
-
simple path diagram

-
a more descriptive diagram
Adding provenance to your data package
Provenance can be added to a data package using datapack and the function in the example code below. But see the vignette or the function documentation for more details.
Example code for provenance:
> dp <- describeWorkflow(dp, sources = doIn, program = progObj, derivations = doOut)
Closing thoughts
Starting to document your data at the beginning of your project will save you time and headache at the end of your project.
Integrating data documentation and publication into your research workflow will increase collaboration efficiency, reproducibility, and impact in the science community.
Exercises
Exercise 1
ORCiDs identify you (like a DOI identifies a paper) and link you to your publications and research products.
If you don’t have an ORCiD, register and obtain your number at https://orcid.org.
Exercise 2
The DataONE federation of repositories requires a unique authentication token for you to upload a data package.
Follow the steps here to obtain your token.
Exercise 3
There are many repositories you could upload your data package to within the DataONE federation.
Pick a repository from the list (located at data/Nodes.csv) and upload a test data package to the “STAGING” environment.
Note: Staging environments require a different authentication token than production environments. See the section on token in the publishing part of the lesson for more details.
Use the example code from the lesson.
Exercise 4
DOIs (digital object identifiers) are almost univerally used to identify papers. They also improve citation and discoverability for your published data packages.
Get a DOI for your published data package.
Use the example code from the lesson.
Exercise 5
If you’re using a data repository to manage versions of your derived data files, you will likely want to update your data file at some point. Use the vignette from the dataone package to “replace an older version of your derived data” with a newer version.
Exercise 6
Provenance is an important description of how your data have been obtained, cleaned, processed, and analyzed. It is being incorporated into descriptions of data packages in some repositories.
Use the describeWorkflow() function from the datapack package to add provenance to your test data package.
Use the example code from the lesson.
If you need to catch-up before a section of code will work, just squish it's 🍅 to copy code above it into your clipboard. Then paste into your interpreter's console, run, and you'll be ready to start in on that section. Code copied by both 🍅 and 📋 will also appear below, where you can edit first, and then copy, paste, and run again.
# Nothing here yet!