Introduction to Land Change Modelling
Lesson 2 with Benoit Parmentier
Contents
#################################### Land Use and Land Cover Change #######################################
############################ Analyze Land Cover change in Houston #######################################
#This script performs analyses for the Exercise 4 of the Geospatial Short Course using aggregated NLCD values.
#The goal is to assess land cover change using two land cover maps in the Houston areas.
#Additional datasets are provided for the land cover change modeling. A model is built for Harris county.
#
#AUTHORS: Benoit Parmentier
#DATE CREATED: 03/16/2018
#DATE MODIFIED: 03/28/2018
#Version: 1
#PROJECT: SESYNC and AAG 2018 Geospatial Short Course
#TO DO:
#
#COMMIT: clean up code for workshop
#
#################################################################################################
###Loading R library and packages
library(sp) # spatial/geographfic objects and functions
library(rgdal) #GDAL/OGR binding for R with functionalities
rgdal: version: 1.2-18, (SVN revision 718)
Geospatial Data Abstraction Library extensions to R successfully loaded
Loaded GDAL runtime: GDAL 2.1.3, released 2017/20/01
Path to GDAL shared files: /usr/share/gdal/2.1
GDAL binary built with GEOS: TRUE
Loaded PROJ.4 runtime: Rel. 4.9.2, 08 September 2015, [PJ_VERSION: 492]
Path to PROJ.4 shared files: (autodetected)
Linking to sp version: 1.2-7
library(spdep) #spatial analyses operations, functions etc.
Loading required package: Matrix
Loading required package: spData
To access larger datasets in this package, install the spDataLarge
package with: `install.packages('spDataLarge',
repos='https://nowosad.github.io/drat/', type='source'))`
library(gtools) # contains mixsort and other useful functions
library(maptools) # tools to manipulate spatial data
Checking rgeos availability: TRUE
library(parallel) # parallel computation, part of base package no
library(rasterVis) # raster visualization operations
Loading required package: raster
Loading required package: lattice
Loading required package: latticeExtra
Loading required package: RColorBrewer
library(raster) # raster functionalities
library(forecast) #ARIMA forecasting
library(xts) #extension for time series object and analyses
Loading required package: zoo
Attaching package: 'zoo'
The following objects are masked from 'package:base':
as.Date, as.Date.numeric
library(zoo) # time series object and analysis
library(lubridate) # dates functionality
Attaching package: 'lubridate'
The following object is masked from 'package:base':
date
library(colorRamps) #contains matlab.like color palette
library(rgeos) #contains topological operations
rgeos version: 0.3-26, (SVN revision 560)
GEOS runtime version: 3.5.1-CAPI-1.9.1 r4246
Linking to sp version: 1.2-5
Polygon checking: TRUE
library(sphet) #contains spreg, spatial regression modeling
Attaching package: 'sphet'
The following object is masked from 'package:raster':
distance
library(BMS) #contains hex2bin and bin2hex, Bayesian methods
library(bitops) # function for bitwise operations
library(foreign) # import datasets from SAS, spss, stata and other sources
#library(gdata) #read xls, dbf etc., not recently updated but useful
library(classInt) #methods to generate class limits
library(plyr) #data wrangling: various operations for splitting, combining data
Attaching package: 'plyr'
The following object is masked from 'package:lubridate':
here
#library(gstat) #spatial interpolation and kriging methods
library(readxl) #functionalities to read in excel type data
library(psych) #pca/eigenvector decomposition functionalities
Attaching package: 'psych'
The following object is masked from 'package:gtools':
logit
library(sf) #spatial objects and functionalities
Linking to GEOS 3.5.1, GDAL 2.1.3, proj.4 4.9.2
library(plotrix) #various graphic functions e.g. draw.circle
Attaching package: 'plotrix'
The following object is masked from 'package:psych':
rescale
library(TOC) # TOC and ROC for raster images
Loading required package: bit
Attaching package bit
package:bit (c) 2008-2012 Jens Oehlschlaegel (GPL-2)
creators: bit bitwhich
coercion: as.logical as.integer as.bit as.bitwhich which
operator: ! & | xor != ==
querying: print length any all min max range sum summary
bit access: length<- [ [<- [[ [[<-
for more help type ?bit
Attaching package: 'bit'
The following object is masked _by_ '.GlobalEnv':
chunk
The following object is masked from 'package:psych':
keysort
The following object is masked from 'package:base':
xor
library(ROCR) # ROCR general for data.frame
Loading required package: gplots
Attaching package: 'gplots'
The following object is masked from 'package:plotrix':
plotCI
The following object is masked from 'package:stats':
lowess
###### Functions used in this script
create_dir_fun <- function(outDir,out_suffix=NULL){
#if out_suffix is not null then append out_suffix string
if(!is.null(out_suffix)){
out_name <- paste("output_",out_suffix,sep="")
outDir <- file.path(outDir,out_name)
}
#create if does not exists
if(!file.exists(outDir)){
dir.create(outDir)
}
return(outDir)
}
##### Parameters and argument set up ###########
#Separate inputs and outputs directories
in_dir_var <- "data"
out_dir <- "."
### General parameters
#NLCD coordinate reference system: we will use this projection rather than TX.
CRS_reg <- "+proj=aea +lat_1=29.5 +lat_2=45.5 +lat_0=23 +lon_0=-96 +x_0=0 +y_0=0 +ellps=GRS80 +towgs84=0,0,0,0,0,0,0 +units=m +no_defs"
file_format <- ".tif" #raster output format
NA_flag_val <- -9999 # NA value assigned to output raster
out_suffix <-"lesson2" #output suffix for the files and ouptu folder #PARAM 8
create_out_dir_param=FALSE # if TRUE, a output dir using output suffix will be created
method_proj_val <- "bilinear" # method option for the reprojection and resampling
gdal_installed <- TRUE #if TRUE, GDAL is used to generate distance files
### Input data files
rastername_county_harris <- "harris_county_mask.tif" #Region of interest: extent of Harris County
elevation_fname <- "srtm_Houston_area_90m.tif" #SRTM elevation
roads_fname <- "r_roads_Harris.tif" #Road count for Harris county
### Aggreagate NLCD input files
infile_land_cover_date1 <- "agg_3_r_nlcd2001_Houston.tif"
infile_land_cover_date2 <- "agg_3_r_nlcd2006_Houston.tif"
infile_land_cover_date3 <- "agg_3_r_nlcd2011_Houston.tif"
infile_name_nlcd_legend <- "nlcd_legend.txt"
infile_name_nlcd_classification_system <- "classification_system_nlcd_legend.xlsx"
######################### START SCRIPT ###############################
## First create an output directory to separate inputs and outputs
if(is.null(out_dir)){
out_dir <- dirname(in_dir) #output will be created in the input dir
}
out_suffix_s <- out_suffix #can modify name of output suffix
if(create_out_dir_param==TRUE){
out_dir <- create_dir_fun(out_dir,out_suffix_s)
setwd(out_dir)
}else{
setwd(out_dir) #use previoulsy defined directory
}
###########################################
### PART I: READ AND VISUALIZE DATA #######
r_lc_date1 <- raster(file.path(in_dir_var,infile_land_cover_date1)) #NLCD 2001
r_lc_date2 <- raster(file.path(in_dir_var,infile_land_cover_date2)) #NLCD 2006
r_lc_date3 <- raster(file.path(in_dir_var,infile_land_cover_date2)) #NLCD 2011
lc_legend_df <- read.table(file.path(in_dir_var,infile_name_nlcd_legend),
stringsAsFactors = F,
sep=",")
head(lc_legend_df) # Inspect data
ID COUNT Red Green Blue NLCD.2006.Land.Cover.Class Opacity
1 0 7854240512 0 0 0 Unclassified 255
2 1 0 0 249 0 255
3 2 0 0 0 0 255
4 3 0 0 0 0 255
5 4 0 0 0 0 255
6 5 0 0 0 0 255
plot(r_lc_date2) # View NLCD 2006, we will need to add the legend use the appropriate palette!!
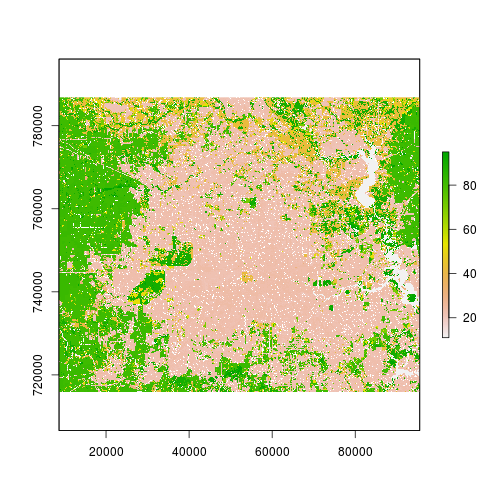
### Let's add legend and examine existing land cover categories
freq_tb_date2 <- freq(r_lc_date2)
head(freq_tb_date2) #view first 5 rows, note this is a matrix object.
value count
[1,] 11 18404
[2,] 21 95347
[3,] 22 93687
[4,] 23 122911
[5,] 24 53352
[6,] 31 6097
### Let's generate a palette from the NLCD legend information to view the existing land cover for 2006.
names(lc_legend_df)
[1] "ID" "COUNT"
[3] "Red" "Green"
[5] "Blue" "NLCD.2006.Land.Cover.Class"
[7] "Opacity"
dim(lc_legend_df) #contains a lot of empty rows
[1] 256 7
lc_legend_df<- subset(lc_legend_df,COUNT>0) #subset the data to remove unsured rows
### Generate a palette color from the input Red, Green and Blue information using RGB encoding:
lc_legend_df$rgb <- paste(lc_legend_df$Red,lc_legend_df$Green,lc_legend_df$Blue,sep=",") #combine
### row 2 correspond to the "open water" category
color_val_water <- rgb(lc_legend_df$Red[2],lc_legend_df$Green[2],lc_legend_df$Blue[2],maxColorValue = 255)
color_val_developed_high <- rgb(lc_legend_df$Red[7],lc_legend_df$Green[7],lc_legend_df$Blue[7],maxColorValue = 255)
lc_col_palette <- c(color_val_water,color_val_developed_high)
barplot(c(1,1),
col=lc_col_palette,
main="Visualization of color palette for NLCD land cover",
names.arg=c("Open water", "Developed, High Intensity"),las=1)
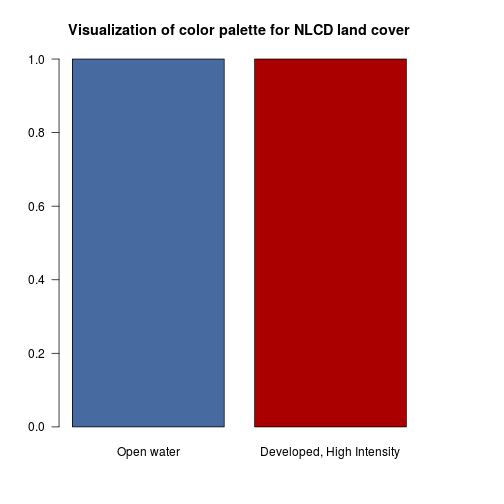
### Let's generate a color for all the land cover categories by using lapply and function
n_cat <- nrow(lc_legend_df)
lc_col_palette <- lapply(1:n_cat,
FUN=function(i){rgb(lc_legend_df$Red[i],lc_legend_df$Green[i],lc_legend_df$Blue[i],maxColorValue = 255)})
lc_col_palette <- unlist(lc_col_palette)
lc_legend_df$palette <- lc_col_palette
r_lc_date2 <- ratify(r_lc_date2) # create a raster layer with categorical information
rat <- levels(r_lc_date2)[[1]] #This is a data.frame with the categories present in the raster
lc_legend_df_date2 <- subset(lc_legend_df,lc_legend_df$ID%in% (rat[,1])) #find the land cover types present in date 2 (2006)
rat$legend <- lc_legend_df_date2$NLCD.2006.Land.Cover.Class #assign it back in case it is missing
levels(r_lc_date2) <- rat #add the information to the raster layer
### Now generate a plot of land cover with the NLCD legend and palette
levelplot(r_lc_date2,
col.regions = lc_legend_df_date2$palette,
scales=list(draw=FALSE),
main = "NLCD 2006")
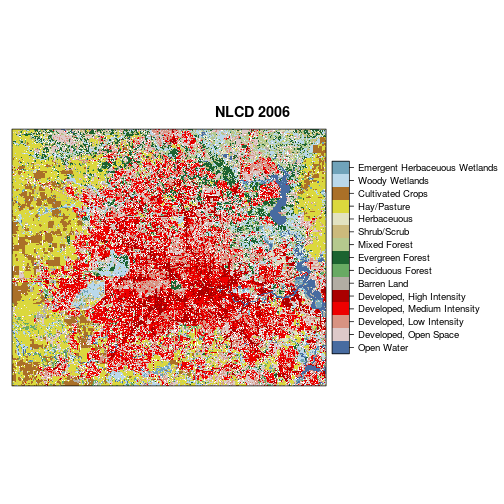
################################################
### PART II : Analyze change and transitions
## As the plot shows for 2006, we have 15 land cover types. Analyzing such complex categories in terms of decreasse (loss), increase (gain),
# persistence in land cover will generate a large number of transitions (potential up to 15*15=225 transitions in this case!)
## To generalize the information, let's aggregate leveraging the hierachical nature of NLCD Anderson Classification system.
lc_system_nlcd_df <- read_xlsx(file.path(in_dir_var,infile_name_nlcd_classification_system))
head(lc_system_nlcd_df) #inspect data
# A tibble: 6 x 5
id_l1 id_l2 name_l1 name_l2 `Classification Descr~
<dbl> <dbl> <chr> <chr> <chr>
1 1. 11. Water Open Water Open Water- areas of ~
2 1. 12. Water Perennial Ice/Snow Perennial Ice/Snow- a~
3 2. 21. Developed Developed, Open Space Developed, Open Space~
4 2. 22. Developed Developed, Low Intensity Developed, Low Intens~
5 2. 23. Developed Developed, Medium Intensity Developed, Medium Int~
6 2. 24. Developed Developed High Intensity Developed High Intens~
### Let's identify existing cover and compute change:
r_stack_nlcd <- stack(r_lc_date1,r_lc_date2)
freq_tb_nlcd <- as.data.frame(freq(r_stack_nlcd,merge=T))
head(freq_tb_nlcd)
value agg_3_r_nlcd2001_Houston agg_3_r_nlcd2006_Houston
1 11 17800 18404
2 21 92135 95347
3 22 89035 93687
4 23 102053 122911
5 24 47830 53352
6 31 4540 6097
dim(lc_system_nlcd_df) # We have categories that are not relevant to the study area and time period.
[1] 20 5
lc_system_nlcd_df <- subset(lc_system_nlcd_df,id_l2%in%freq_tb_nlcd$value )
dim(lc_system_nlcd_df) # Now 15 land categories instead of 20.
[1] 15 5
### Selectet relevant columns for the reclassification
rec_df <- lc_system_nlcd_df[,c(2,1)]
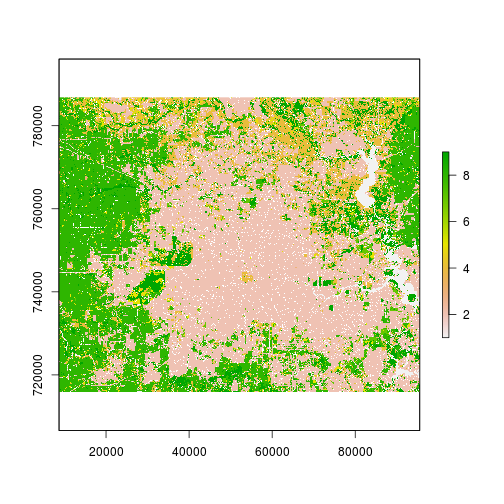
r_date1_rec <- subs(r_lc_date1,rec_df,by="id_l2","id_l1")
r_date2_rec <- subs(r_lc_date2,rec_df,by="id_l2","id_l1")
plot(r_date1_rec)
rec_xtab_df <- crosstab(r_date1_rec,r_date2_rec,long=T)
names(rec_xtab_df) <- c("2001","2011","freq")
head(rec_xtab_df)
2001 2011 freq
1 1 1 17285
2 2 1 7
3 3 1 231
4 4 1 76
5 5 1 21
6 7 1 73
dim(rec_xtab_df) #9*9 possible transitions if we include NA values
[1] 81 3
print(rec_xtab_df) # View the full table
2001 2011 freq
1 1 1 17285
2 2 1 7
3 3 1 231
4 4 1 76
5 5 1 21
6 7 1 73
7 8 1 306
8 9 1 302
9 <NA> 1 103
10 1 2 74
11 2 2 325161
12 3 2 1111
13 4 2 10115
14 5 2 1364
15 7 2 3243
16 8 2 12540
17 9 2 3987
18 <NA> 2 7702
19 1 3 176
20 2 3 10
21 3 3 2815
22 4 3 386
23 5 3 232
24 7 3 145
25 8 3 1929
26 9 3 314
27 <NA> 3 90
28 1 4 8
29 2 4 40
30 3 4 10
31 4 4 77877
32 5 4 192
33 7 4 27
34 8 4 48
35 9 4 41
36 <NA> 4 242
37 1 5 34
38 2 5 14
39 3 5 70
40 4 5 807
41 5 5 13782
42 7 5 752
43 8 5 264
44 9 5 87
45 <NA> 5 186
46 1 7 95
47 2 7 32
48 3 7 115
49 4 7 996
50 5 7 452
51 7 7 16967
52 8 7 411
53 9 7 91
54 <NA> 7 213
55 1 8 3
56 2 8 17
57 3 8 7
58 4 8 127
59 5 8 37
60 7 8 14
61 8 8 154887
62 9 8 110
63 <NA> 8 83
64 1 9 38
65 2 9 19
66 3 9 51
67 4 9 116
68 5 9 11
69 7 9 19
70 8 9 52
71 9 9 58478
72 <NA> 9 145
73 1 <NA> 87
74 2 <NA> 5753
75 3 <NA> 130
76 4 <NA> 1654
77 5 <NA> 271
78 7 <NA> 516
79 8 <NA> 1673
80 9 <NA> 691
81 <NA> <NA> 32745
which.max(rec_xtab_df$freq)
[1] 11
rec_xtab_df[11,] # Note the most important transition is persistence!!
2001 2011 freq
11 2 2 325161
### Let's rank the transition:
class(rec_xtab_df)
[1] "data.frame"
is.na(rec_xtab_df$freq)
[1] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[12] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[23] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[34] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[45] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[56] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[67] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[78] FALSE FALSE FALSE FALSE
rec_xtab_df_ranked <- rec_xtab_df[order(rec_xtab_df$freq,decreasing=T) , ]
head(rec_xtab_df_ranked) # Unsurprsingly, top transitions are persistence categories
2001 2011 freq
11 2 2 325161
61 8 8 154887
31 4 4 77877
71 9 9 58478
81 <NA> <NA> 32745
1 1 1 17285
### Let's examine the overall change in categories rather than transitions
label_legend_df <- data.frame(ID=lc_system_nlcd_df$id_l1,name=lc_system_nlcd_df$name_l1)
r_stack <- stack(r_date1_rec,r_date2_rec)
lc_df <- freq(r_stack,merge=T)
names(lc_df) <- c("value","date1","date2")
lc_df$diff <- lc_df$date2 - lc_df$date1 #difference for each land cover categories over the 2001-2011 time period
head(lc_df) # Quickly examine the output
value date1 date2 diff
1 1 17800 18404 604
2 2 331053 365297 34244
3 3 4540 6097 1557
4 4 92154 78485 -13669
5 5 16362 15996 -366
6 7 21756 19372 -2384
### Add relevant categories
lc_df <- merge(lc_df,label_legend_df,by.x="value",by.y="ID",all.y=F)
lc_df <- lc_df[!duplicated(lc_df),] #remove duplictates
head(lc_df) # Note the overall cahnge
value date1 date2 diff name
1 1 17800 18404 604 Water
2 2 331053 365297 34244 Developed
6 3 4540 6097 1557 Barren
7 4 92154 78485 -13669 Forest
10 5 16362 15996 -366 Shrubland
11 7 21756 19372 -2384 Herbaceous
#### Now visualize the overall land cover changes
barplot(lc_df$diff,names.arg=lc_df$name,las=2)
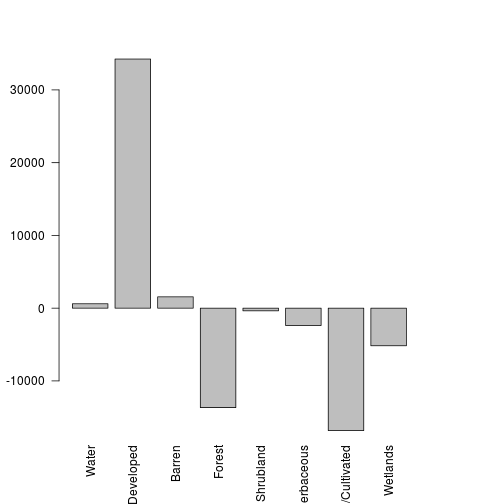
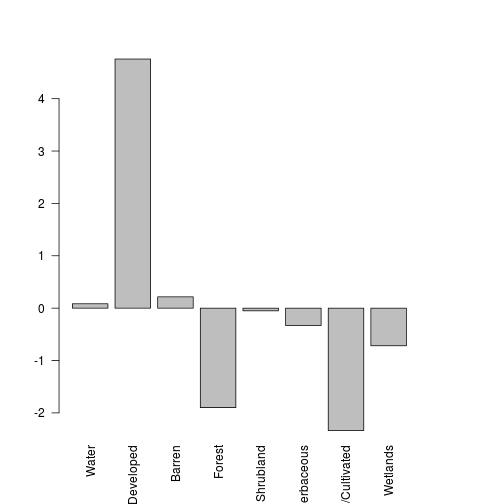
total_val <- sum(lc_df$date1)
lc_df$perc_change <- 100*lc_df$diff/total_val
barplot(lc_df$perc_change,names.arg=lc_df$name,las=2)
### Create a change image to map all pixels that transitioned to the developed category:
r_cat2 <- r_date2_rec==2 # developed on date 2
r_not_cat2 <- r_date1_rec!=2 #remove areas that were already developed in date1, we do not want persistence
r_change <- r_cat2 * r_not_cat2 #mask
plot(r_change,main="Land transitions to developed over 2001-2011")
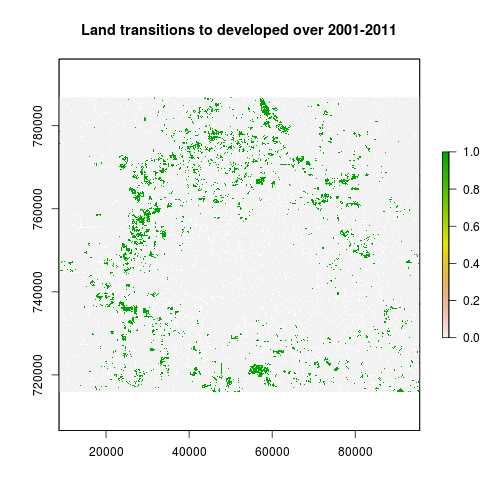
change_tb <- freq(r_change) #Find out how many pixels transitions to developed
#####################################
############# PART III: Process and Prepare variables for land change modeling ##############
## y= 1 if change to urban over 2001-2011
### Explanatory variables:
#var1: distance to existing urban in 2001
#var2: distance to road in 2001
#var3: elevation, low slope better for new development
#var4: past land cover state that may influence future land change
## 1) Generate var1 and var2 : distance to developed and distance to roads
### Distance to existing in 2001: prepare information
r_cat2<- r_date1_rec==2 #developed in 2001
plot(r_cat2)
cat_bool_fname <- "developed_2001.tif" #input for the distance to road computation
writeRaster(r_cat2,filename = cat_bool_fname,overwrite=T)
### Read in data for road count
r_roads <- raster(file.path(in_dir_var,roads_fname))
plot(r_roads,colNA="black")
res(r_roads)
[1] 30 30
#### Aggregate to match the NLCD data resolution
r_roads_90m <- aggregate(r_roads,
fact=3, #factor of aggregation in x and y
fun=mean) #function used in aggregation values
plot(r_roads_90m)
r_roads_bool <- r_roads_90m > 0
plot(r_roads_bool)
roads_bool_fname <- "roads_bool.tif" #input for the distance to road computation
writeRaster(r_roads_bool,filename = roads_bool_fname,overwrite=T)
### This part could be transformed into a function but we keep it for clarity and learning:
if(gdal_installed==TRUE){
## Distance from developed land in 2001
srcfile <- cat_bool_fname
dstfile_developed <- file.path(out_dir,paste("developed_distance_",out_suffix,file_format,sep=""))
n_values <- "1"
### Prepare GDAL command: note that gdal_proximity doesn't like when path is too long
cmd_developed_str <- paste("gdal_proximity.py",
basename(srcfile),
basename(dstfile_developed),
"-values",n_values,sep=" ")
### Distance from roads
srcfile <- roads_bool_fname
dstfile_roads <- file.path(out_dir,paste("roads_bool_distance_",out_suffix,file_format,sep=""))
n_values <- "1"
### Prepare GDAL command: note that gdal_proximity doesn't like when path is too long
cmd_roads_str <- paste("gdal_proximity.py",basename(srcfile),
basename(dstfile_roads),
"-values",n_values,sep=" ")
sys_os <- as.list(Sys.info())$sysname #Find what OS system is in use.
if(sys_os=="Windows"){
shell(cmd_developed_str)
shell(cmd_roads_str)
}else{
system(cmd_developed_str)
system(cmd_roads_str)
}
r_roads_distance <- raster(dstfile_roads)
r_developed_distance <- raster(dstfile_developed)
}else{
r_developed_distance <- raster(file.path(in_dir,paste("developed_distance_",file_format,sep="")))
r_roads_distance <- raster(file.path(in_dir_var,paste("roads_bool_distance",file_format,sep="")))
}
plot(r_developed_distance) #This is at 90m.
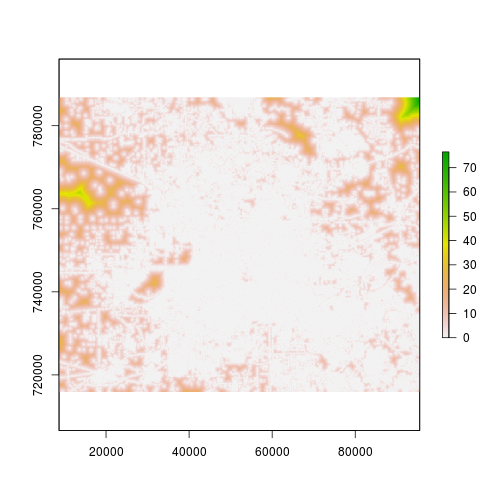
plot(r_roads_distance) #This is at 90m.
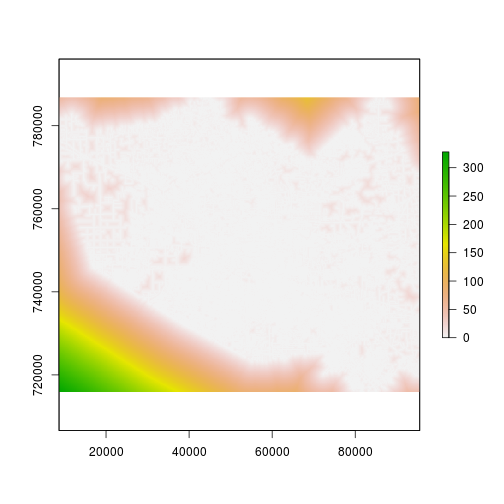
#Now rescale the distance...
min_val <- cellStats(r_roads_distance,min)
max_val <- cellStats(r_roads_distance,max)
r_roads_dist <- (max_val - r_roads_distance) / (max_val - min_val) #high values close to 1 for areas close to roads
min_val <- cellStats(r_developed_distance,min)
max_val <- cellStats(r_developed_distance,max)
r_developed_dist <- (max_val - r_developed_distance) / (max_val - min_val)
## 2) Generate var3 : slope
r_elevation <- raster(file.path(in_dir_var,elevation_fname))
projection(r_elevation) # This is not in the same projection as the study area.
[1] "+proj=longlat +datum=WGS84 +no_defs +ellps=WGS84 +towgs84=0,0,0"
r_elevation_reg <- projectRaster(from= r_elevation, #input raster to reproject
to= r_date1_rec, #raster with desired extent, resolution and projection system
method= method_proj_val) #method used in the reprojection
r_slope <- terrain(r_elevation_reg,unit="degrees")
## 3) Generate var4 : past land cover state
### reclass Land cover
r_mask <- r_date1_rec==2 #Remove developed land from
r_date1_rec_masked <- mask(r_date1_rec,r_mask,maskvalue=1)
plot(r_date1_rec_masked)
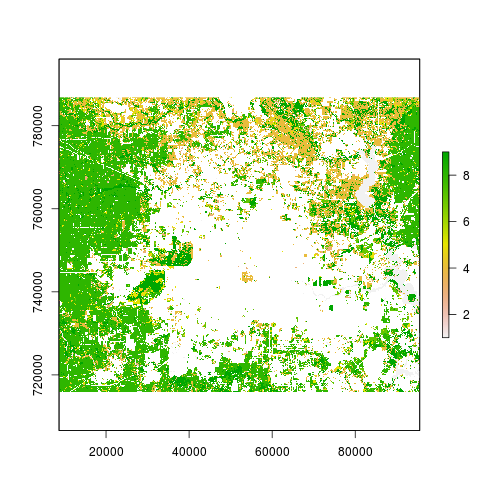
#####################################
############# PART IV: Run Model and perform assessment ##############
##############
###### Step 1: Consistent masking and generate mask removing water (1) and developed (2) in 2001
#r_mask <- (r_date1_rec!=2)*(r_date1_rec!=1)*r_county_harris
r_mask <- (r_date1_rec!=2)*(r_date1_rec!=1)
plot(r_mask)
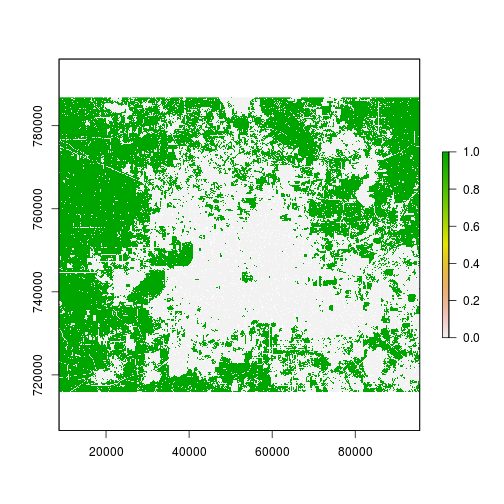
NAvalue(r_mask) <- 0
### Read in focus area for the modeling:
r_county_harris <- raster(file.path(in_dir_var,rastername_county_harris))
### Screen for area of interest
r_mask <- r_mask * r_county_harris
r_mask[r_mask==0]<-NA
plot(r_mask)
### Check the number of NA and pixels in the study area:
tb_study_area <- freq(r_mask)
print(tb_study_area)
value count
[1,] 1 220338
[2,] NA 541047
## Generate dataset for Harris county
r_variables <- stack(r_change,r_date1_rec_masked,r_slope,r_roads_dist,r_developed_dist)
r_variables <- mask(r_variables,mask=r_mask) # mask to keep relevant area
names(r_variables) <- c("change","land_cover","slope","roads_dist","developed_dist")
NAvalue(r_variables) <- NA_flag_val
## Examine all the variables
plot(r_variables)
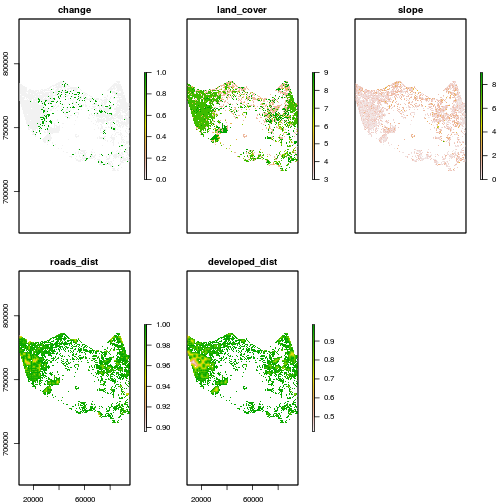
### Check for consistency in mask:
NA_freq_tb <- freq(r_variables,value=NA,merge=T)
### Notice that the number of NA is not consistent.
### Let's recombine all NA for consstencies:
plot(r_variables)
r_NA <- r_variables > -1 #There are no negative values in the raster stack
r_valid_pixels <- overlay(r_NA,fun=sum)
plot(r_valid_pixels)
dim(r_NA)
[1] 789 965 5
freq(r_valid_pixels)
value count
[1,] 5 215437
[2,] NA 545948
r_mask <- r_valid_pixels > 0
r_variables <- mask(r_variables,r_mask)
#r_variables <- freq(r_test2,value=NA,merge=T)
names(r_variables) <- c("change","land_cover","slope","roads_dist","developed_dist")
###############
###### Step 2: Fit glm model and generate predictions
variables_df <- na.omit(as.data.frame(r_variables))
dim(variables_df)
[1] 215437 5
variables_df$land_cover <- as.factor(variables_df$land_cover)
variables_df$change <- as.factor(variables_df$change)
names(variables_df)
[1] "change" "land_cover" "slope" "roads_dist"
[5] "developed_dist"
#names(variables_df) <- c("change","land_cover","elevation","roads_dist","developed_dist")
mod_glm <- glm(change ~ land_cover + slope + roads_dist + developed_dist,
data=variables_df , family=binomial())
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
print(mod_glm)
Call: glm(formula = change ~ land_cover + slope + roads_dist + developed_dist,
family = binomial(), data = variables_df)
Coefficients:
(Intercept) land_cover4 land_cover5 land_cover7
-308.5419 -0.9241 -1.2272 -0.7449
land_cover8 land_cover9 slope roads_dist
-1.0081 -1.2218 -0.2112 298.0062
developed_dist
11.1344
Degrees of Freedom: 215436 Total (i.e. Null); 215428 Residual
Null Deviance: 142300
Residual Deviance: 112900 AIC: 112900
summary(mod_glm)
Call:
glm(formula = change ~ land_cover + slope + roads_dist + developed_dist,
family = binomial(), data = variables_df)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.3712 -0.5328 -0.2546 -0.0418 6.6490
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -308.54187 3.05822 -100.89 <2e-16 ***
land_cover4 -0.92415 0.04763 -19.40 <2e-16 ***
land_cover5 -1.22723 0.06081 -20.18 <2e-16 ***
land_cover7 -0.74491 0.05151 -14.46 <2e-16 ***
land_cover8 -1.00809 0.04851 -20.78 <2e-16 ***
land_cover9 -1.22180 0.05087 -24.02 <2e-16 ***
slope -0.21119 0.01307 -16.16 <2e-16 ***
roads_dist 298.00624 3.07499 96.91 <2e-16 ***
developed_dist 11.13435 0.26236 42.44 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 142342 on 215436 degrees of freedom
Residual deviance: 112871 on 215428 degrees of freedom
AIC: 112889
Number of Fisher Scoring iterations: 8
r_p <- predict(r_variables, mod_glm, type="response")
plot(r_p)
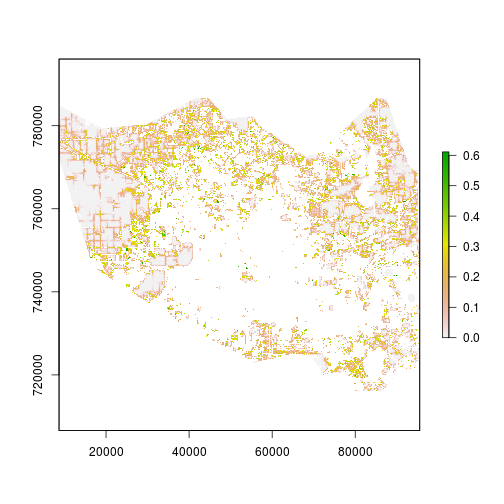
histogram(r_p)
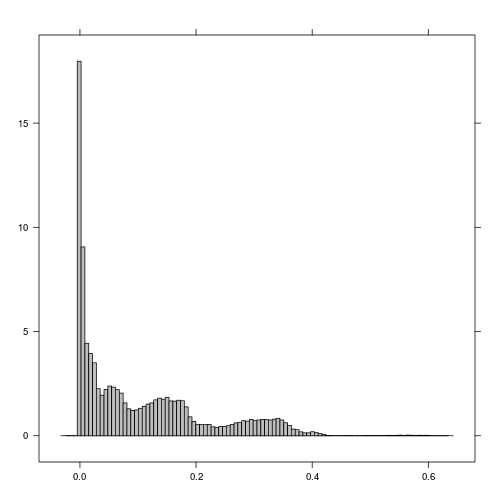
###############
###### Step 3: Model assessment with ROC
## We use the TOC package since it allows for the use of raster layers.
r_change_harris <- subset(r_variables,"change")
## These are the inputs for the assessment
plot(r_change_harris) # boolean reference variable
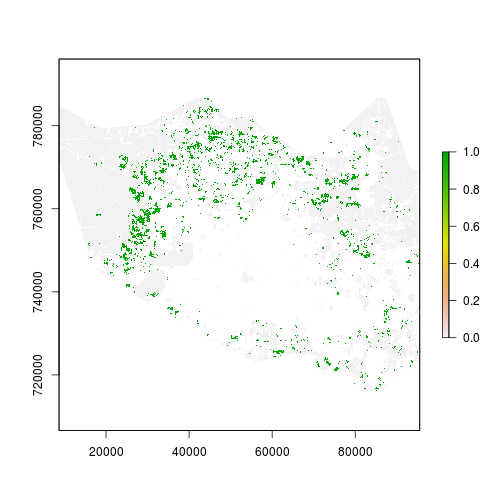
plot(r_mask) # mask for relevant observation
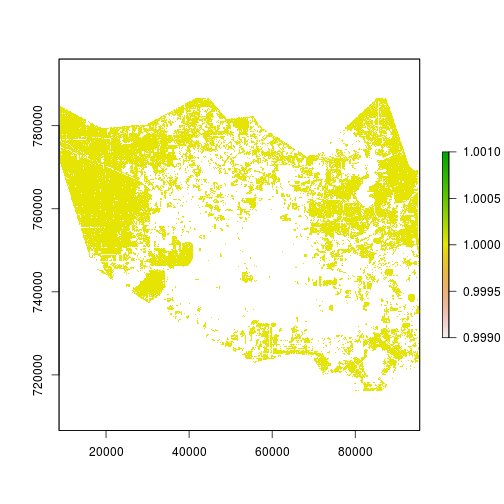
plot(r_p) # index variable to assess
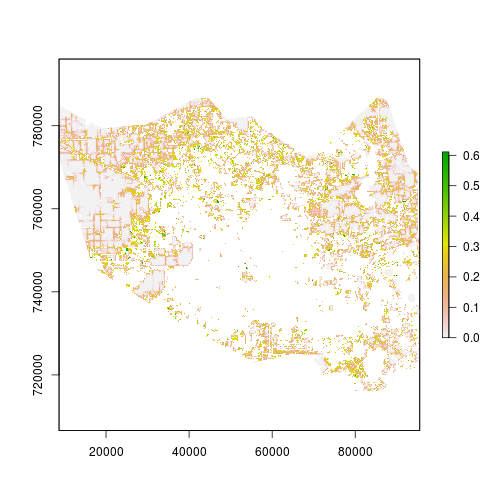
roc_rast <- ROC(index=r_p,
boolean=r_change_harris,
mask=r_mask,
nthres=100)
plot(roc_rast)
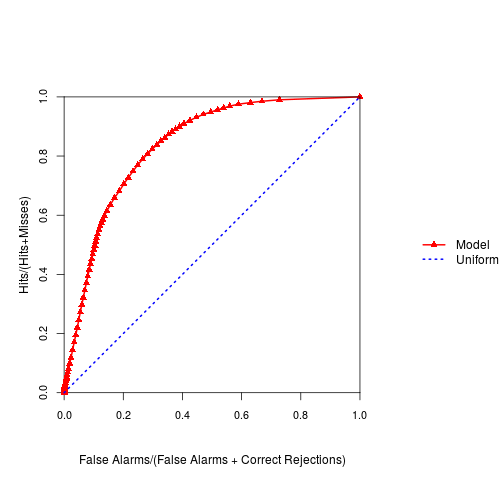
slot(roc_rast,"AUC") #this is the AUC from ROC for the logistic modeling
[1] 0.836335
toc_rast <- TOC(index=r_p,
boolean=r_change_harris,
mask=r_mask,
nthres=100)
plot(toc_rast)
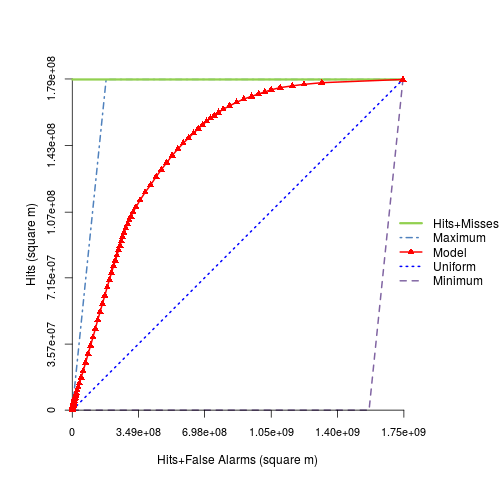
slot(toc_rast,"AUC") #this is the AUC from TOC for the logistic modeling
[1] 0.836335
############################### End of script #####################################