Raster Time-Series
Wildfire in Alaska
Lesson 3 with Ian Carroll
Lesson Objectives
- Observe characteristics of wildfire in remote sensing data
- Find “unusual” pixels in time-series of raster data
Specific Achievements
- Distinguish “stacks” from “bricks”
- Use MODIS derived NDVI
- Execute efficient raster calculations
- Perform PCA on “bricks”
Logistics
Load packages, define global variables, and take care of remaining logistics at the very top.
library(sf)
library(raster)
library(ggplot2)
out <- 'outputs_raster_ts'
dir.create(out, showWarnings = FALSE)
Raster upon Raster
The raster library uniformly handles two-dimensional data as a
RasterLayer object, created with the raster() function. There are several
cases where raster data has a third dimension.
- a multi-band image (e.g. Landsat scenes)
- closely related images (e.g. time series of NDVI values)
- three dimensional data (e.g. GCM products)
Any RasterLayer objects that have the same extent and resolution (i.e. cover
the same space with the same number of rows and columns) can be combined into
one of two container types:
- a
RasterStackcreated withstack() - a
RasterBrickcreated withbrick()
Raster Stacks
The layers of a “stack” can refer to data from separate files, or even a mix of data on disk and data in memory.
ndvi_yrly <- Sys.glob('data/r_ndvi_*.tif')
> ndvi_yrly
[1] "data/r_ndvi_2001_2009_filling6__STA_year2_Amplitude0.tif"
[2] "data/r_ndvi_2001_2009_filling6__STA_year9_Amplitude0.tif"
ndvi <- stack(ndvi_yrly)
names(ndvi) <- c(
'Avg NDVI 2002',
'Avg NDVI 2009')
> plot(ndvi)
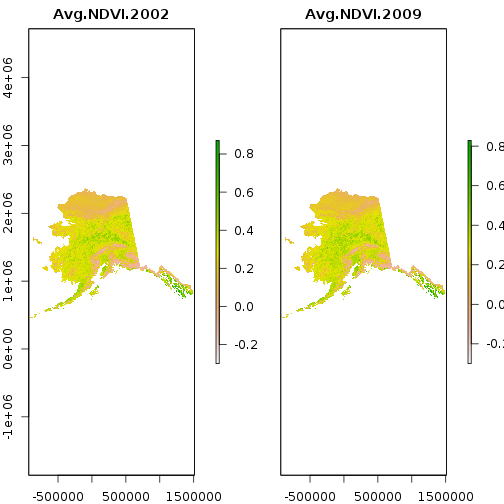
The data source for the first layer is the first file in the list. Note that the CRS is missing: it is quite possible to stack files with different projections. They only have to share a common extent and resolution.
> raster(ndvi, 1)
class : RasterLayer
dimensions : 1951, 2441, 4762391 (nrow, ncol, ncell)
resolution : 1000.045, 999.9567 (x, y)
extent : -930708.7, 1510401, 454027.3, 2404943 (xmin, xmax, ymin, ymax)
coord. ref. : NA
data source : /nfs/public-data/training/r_ndvi_2001_2009_filling6__STA_year2_Amplitude0.tif
names : Avg.NDVI.2002
values : -0.3, 0.8713216 (min, max)
Set the CRS using the EPSG code 3338 for an Albers Equal Area projection of Alaska using the NAD38 datum.
crs(ndvi) <- '+init=epsg:3338'
> raster(ndvi, 0)
class : RasterLayer
dimensions : 1951, 2441, 4762391 (nrow, ncol, ncell)
resolution : 1000.045, 999.9567 (x, y)
extent : -930708.7, 1510401, 454027.3, 2404943 (xmin, xmax, ymin, ymax)
coord. ref. : +init=epsg:3338 +proj=aea +lat_1=55 +lat_2=65 +lat_0=50 +lon_0=-154 +x_0=0 +y_0=0 +datum=NAD83 +units=m +no_defs +ellps=GRS80 +towgs84=0,0,0
The ndvi object only takes up a tiny amount of memory. The pixel values
take up much more space on disk, as you can see in the file browser.
> print(object.size(ndvi),
+ units = 'KB',
+ standard = 'SI')
29.6 kB
Why so small? The ndvi object is only metadata and a pointer to where the
pixel values are saved on disk.
> inMemory(ndvi)
[1] FALSE
Whereas read.csv() would load the named file into memory, the
raster library handles files like a database where possible. The
values can be accessed, to make those plots for example, but are not held in
memory. This is the key to working with deep stacks of large rasters.
Wildfires in Alaska
Many and massive wildfires burned in Alaska and the Yukon between 2001 and 2009. Three large fires that burned during this period (their locations are in a shapefile) occured within boreal forest areas of central Alaska.
scar <- read_sf(
'data/OVERLAY_ID_83_399_144_TEST_BURNT_83_144_399_reclassed',
crs = 3338)
plot(ndvi[[1]])
plot(st_geometry(scar), add = TRUE)
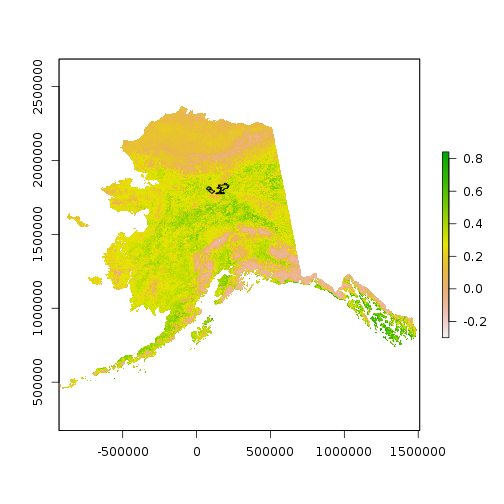
For faster processing in this lesson, crop the NDVI imagery to the smaller extent of the shapefile.
burn_bbox <-
extent(matrix(st_bbox(scar), 2))
ndvi <- crop(ndvi, burn_bbox)
> plot(ndvi[[1]], ext = burn_bbox)
> plot(st_geometry(scar), add = TRUE)
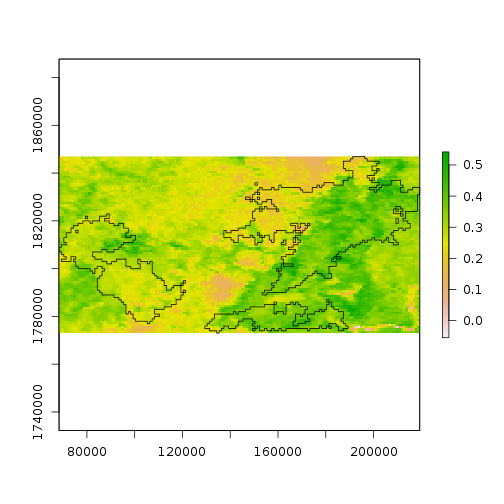
Notice, however, that the NDVI values are now stored in memory. That’s okay for this part of the lesson; we’ll see the alternative shortly.
> inMemory(ndvi)
[1] TRUE
Pixel Change
Use element-wise subtration to give a difference raster, where negative values indicate a higher NDVI in 2002, or a decrease in NDVI from 2002 to 2009.
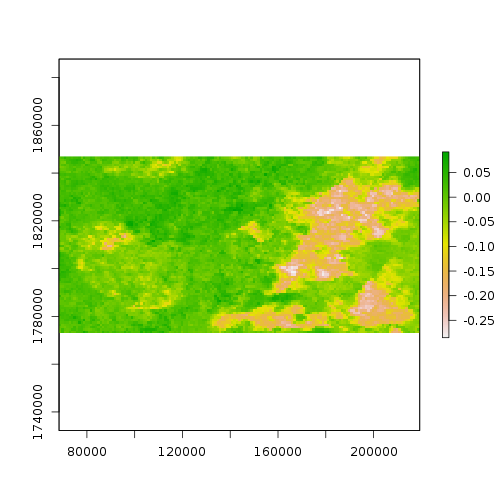
diff_ndvi <- ndvi[[2]] - ndvi[[1]]
names(diff_ndvi) <- 'Difference'
> plot(diff_ndvi)
The histogram shows clearly that change in NDVI within this corner of Alaska clusters around two modes.
> hist(diff_ndvi)
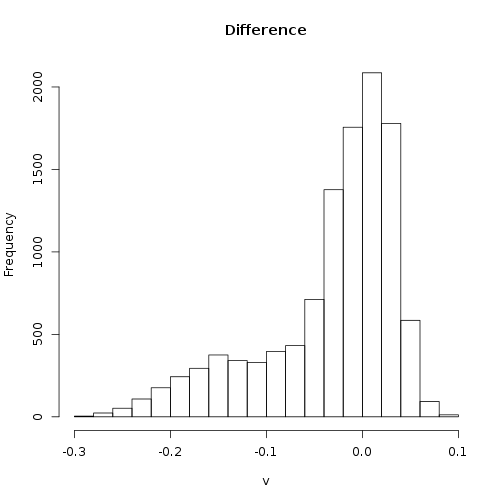
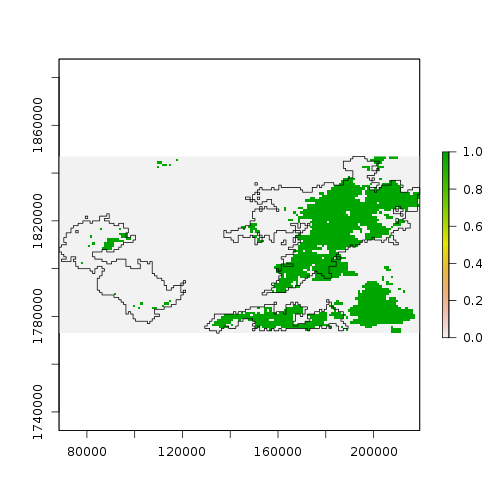
One way to “classify” pixels as potentially affected by wildfire is to threshold the differrence. Pixels below “-0.1” mostly belong to the smaller mode, and may represent impacts of wildfire.
> plot(diff_ndvi < -0.1)
> plot(st_geometry(scar), add = TRUE)
The threshold value could also be defined in terms of the variability among pixels.
Centering and scaling the pixel values requires computation of their
mean and standard variation. The cellStats function efficiently applies a few
common functions across large rasters, regardless of whether the values are in
memory or on disk.
diff_ndvi_mean <-
cellStats(diff_ndvi, 'mean')
diff_ndvi_sd <-
cellStats(diff_ndvi, 'sd')
Mathematical operations with rasters and scalars work as expected; scalar values are repeated for each cell in the array.
The difference threshold of “-0.1” appears roughly equivalent to a threshold of 1 standard deviation below zero.
diff_ndvi_stdz <-
(diff_ndvi - diff_ndvi_mean) /
diff_ndvi_sd
names(diff_ndvi_stdz) <- 'Std. Diff.'
> hist(diff_ndvi_stdz, breaks = 20)
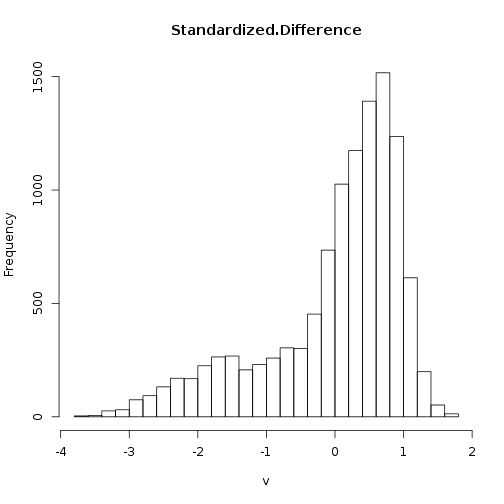
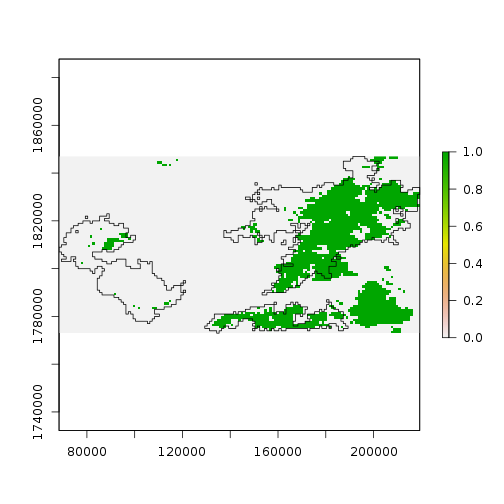
Standardizing the pixel values does not change the overal result.
> plot(diff_ndvi_stdz < -1)
> plot(st_geometry(scar), add = TRUE)
Raster Time Series
Take a closer look at NDVI using products covering 16 day periods in 2005. These images are stored as separate files on disk, all having the same extent and resolution.
ndvi_16day <- Sys.glob(
'data/NDVI_alaska_2005/*.tif')
ndvi <- stack(ndvi_16day)
crs(ndvi) <- '+init=epsg:3338'
Lacking other metadata, extract the date of each image from its filename.
dates <- as.Date(
sub('alaska_NDVI_', '', names(ndvi)),
'%Y_%m_%d')
names(ndvi) <- format(dates, '%b %d %Y')
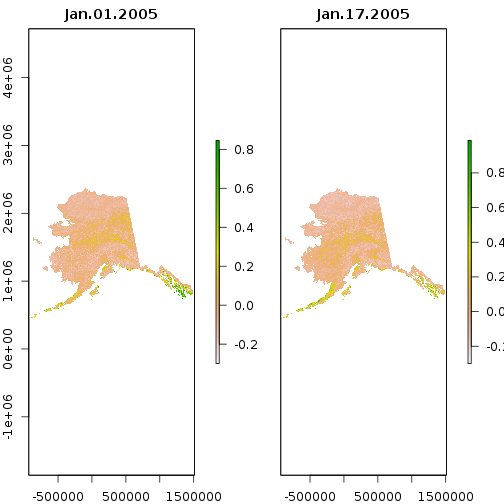
> plot(subset(ndvi, 1:2))
Raster Bricks
A RasterBrick representation of tightly integrated raster layers, such as a
time series of remote sensing data from sequential overflights, has advantages
for speed but limitations on flexibility.
A RasterStack is more flexible because it can mix values stored on disk with
those in memory. Adding a layer of in-memory values to a RasterBrick causes the
entire brick to be loaded into memory, which may not be possible given the
available memory.
For training purposes, again crop the NDVI data to the bounding box of the wildfires shapefile. But this time, avoid reading the values into memory.
ndvi <- crop(ndvi, burn_bbox,
filename = file.path(out, 'crop_alaska_ndvi.grd'),
overwrite = TRUE)
Notice that crop creates a RasterBrick. In fact, we have been working with a
RasterBrick in memory since first using crop.
> ndvi
class : RasterBrick
dimensions : 74, 151, 11174, 23 (nrow, ncol, ncell, nlayers)
resolution : 1000.045, 999.9566 (x, y)
extent : 68336.16, 219342.9, 1772970, 1846967 (xmin, xmax, ymin, ymax)
coord. ref. : +init=epsg:3338 +proj=aea +lat_1=55 +lat_2=65 +lat_0=50 +lon_0=-154 +x_0=0 +y_0=0 +datum=NAD83 +units=m +no_defs +ellps=GRS80 +towgs84=0,0,0
data source : /home/ian/training/handouts/build/raster-time-series-alaska-lesson/outputs_raster_ts/crop_alaska_ndvi.grd
names : Jan.01.2005, Jan.17.2005, Feb.02.2005, Feb.18.2005, Mar.06.2005, Mar.22.2005, Apr.07.2005, Apr.23.2005, May.09.2005, May.25.2005, Jun.10.2005, Jun.26.2005, Jul.12.2005, Jul.28.2005, Aug.13.2005, ...
min values : -0.19518, -0.20000, -0.19900, -0.18050, -0.12120, -0.09540, -0.03910, -0.11290, -0.09390, -0.15520, -0.18400, -0.16780, -0.18000, -0.17200, -0.18600, ...
max values : 0.3337000, 0.3633000, 0.4949000, 0.3514000, 0.4898000, 0.7274000, 0.6268000, 0.5879000, 0.9076000, 0.9190000, 0.8807000, 0.9625000, 0.8810000, 0.9244000, 0.9493000, ...
So Many Data
The immediate challenge is trying to represent the data in ways we can explore and interpret the characteristics of wildfire visible by remote sensing.
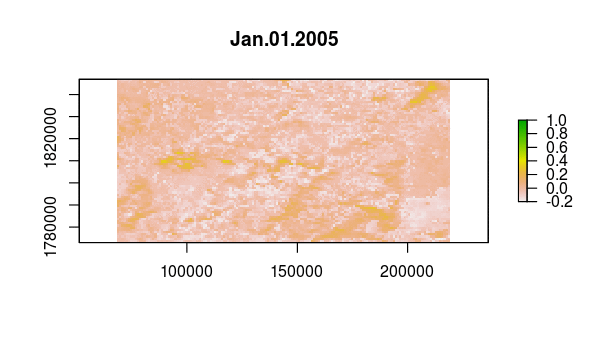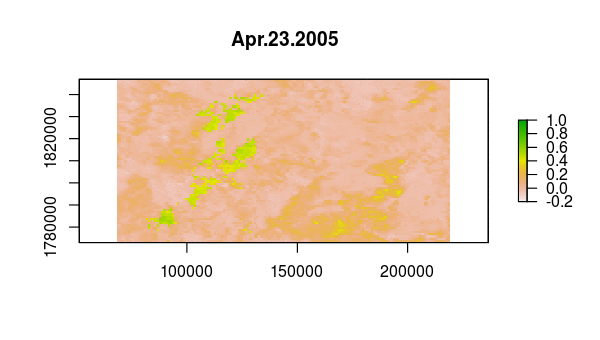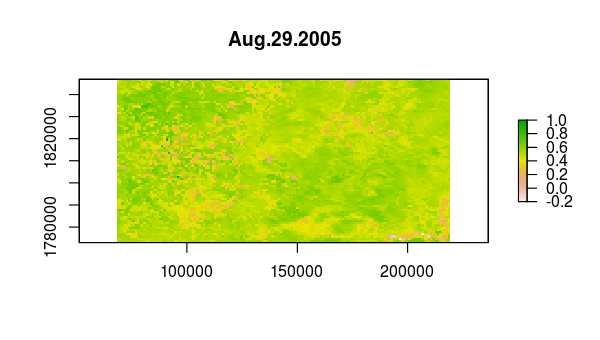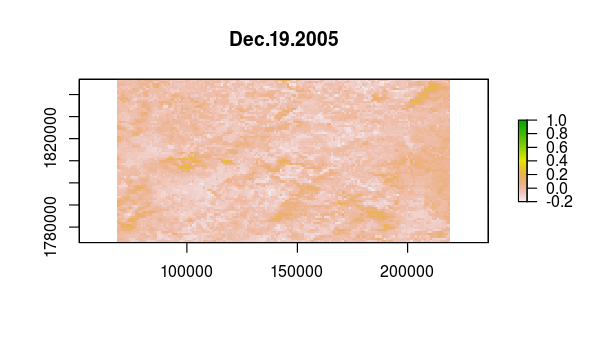
> animate(ndvi, pause = 0.5, n = 1)
Pixel Time Series
Verify that something happened very abruptly (fire!?) by plotting the time series at pixels corresponding to locations with dramatic NDVI variation in the layer from Aug 13, 2005.
idx <- match('Aug.13.2005', names(ndvi))
plot(ndvi[[idx]])
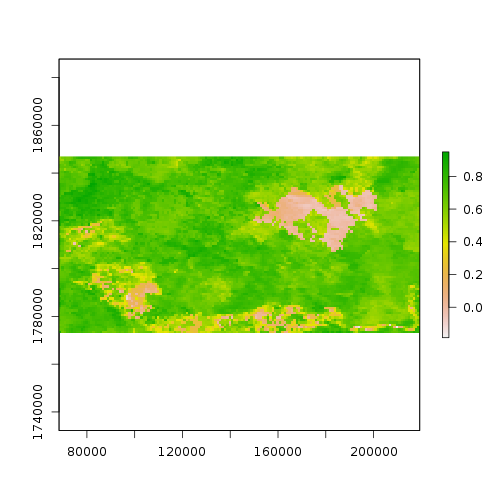
> pixel <- click(ndvi[[idx]], cell = TRUE)
> pixel
“Hard code” these pixel values into your worksheet
pixel <- c(2813, 3720, 2823, 4195, 9910)
scar_pixel <- data.frame(
Date = rep(dates, each = length(pixel)),
cell = rep(pixel, length(dates)),
Type = 'burn scar?',
NDVI = c(ndvi[pixel]))
Repeat the selection with click for “normal” looking pixels.
pixel <- c(1710, 4736, 7374, 1957, 750)
normal_pixel <- data.frame(
Date = rep(dates, each = length(pixel)),
cell = rep(pixel, length(dates)),
Type = 'normal',
NDVI = c(ndvi[pixel]))
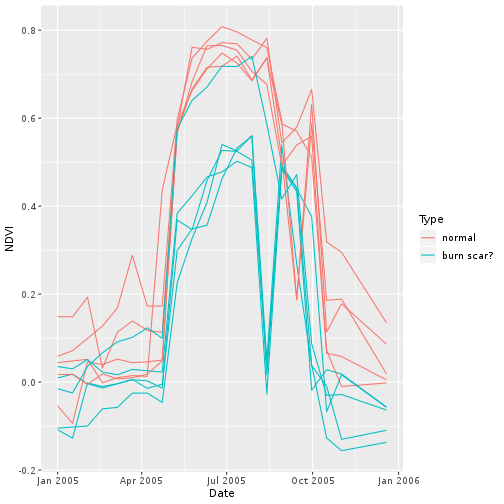
Join your haphazard samples together for comparison as time series.
pixel <- rbind(normal_pixel, scar_pixel)
ggplot(pixel,
aes(x = Date, y = NDVI,
group = cell, col = Type)) +
geom_line()
Zonal Averages
Cannot very well analyze the time series for every pixel, so we have to reduce the dimensionality of the data. One way is to summarize it by “zones” defined by another spatial data source.
> ?zonal
Currently we have raster data (ndvi) and vector data (scar). In order to
aggregate by polygon, we have to join these two datasets. There are two
approaches. 1) Treat the raster data as POINT geometries in a table and perform
a spatial join to the table with POLYGON geometries. 2) Turn the polygons into a
raster and summarize the raster masked for each polygon. Let’s persue option 2,
but take a shortcut due to the presence of invalid geometries in the shapefile.
Typically we could convert simple features to raster with the rasterize
function, but not all these geometries are well defined.
# Does not work, due to invalid geometries.
scar_geom <-
as(st_geometry(scar), 'Spatial')
scar_zone <- rasterize(scar_geom, ndvi,
background = 0,
filename = 'results/scar.grd',
overwrite = TRUE)
Fortunately, we have the rasterized version from another source.
scar_zone <- raster('data/r_OVERLAY_ID_83_399_144_TEST_BURNT_83_144_399_reclassed.tif')
crs(scar_zone) <- '+init=epsg:3338'
scar_zone <- crop(scar_zone, ndvi)
The zonal function calculates fun over each zone
scar_ndvi <-
zonal(ndvi, scar_zone, "mean")
Rearrange the data for vizualization as a time series.
zone <- factor(scar_ndvi[, 1])
scar_ndvi <- scar_ndvi[, -1]
scar_zone <- data.frame(
Date = rep(dates, each = nrow(scar_ndvi)),
Zone = rep(zone, length(dates)),
NDVI = c(scar_ndvi))
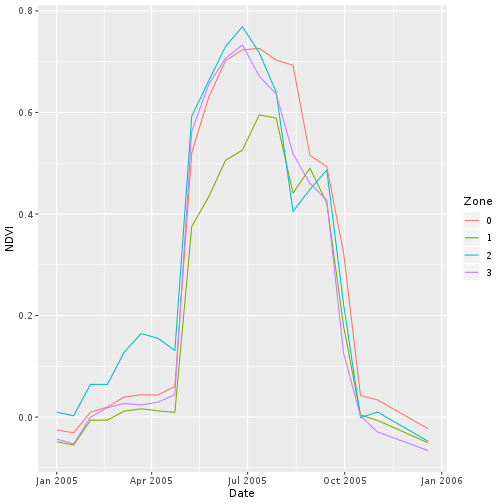
What appears to be the most pronounced signal in this view is an early loss of greenness compared to the background NDVI.
> ggplot(scar_zone,
+ aes(x = Date, y = NDVI,
+ col = Zone)) +
+ geom_line()
Zonal Statistics
- provide a very clean reduction of raster time-series for easy vizualization
- require pre-defined zones!
Eliminating Time
Because changes to NDVI at each pixel follow a similar pattern over the course of a year, the slices are highly correlated. Consider representing the NDVI values as a simple matrix with
- each time slice as a variable
- each pixel as an observation
PCA is a technique for reducing dimensionality of a dataset based on correlation between variables. The method proceeds either by eigenvalue decomposition of a covariance matrix or singular-value decomposition of the entire dataset.
To perform PCA on raster data, it’s efficient to use specialized tools that calculate a covariance matrix without reading in that big data matrix.
ndvi_lS <- layerStats(
ndvi, 'cov', na.rm = TRUE)
ndvi_mean <- ndvi_lS[['mean']]
ndvi_cov <- ndvi_lS[['covariance']]
ndvi_cor <- cov2cor(ndvi_cov)
The layerStats function only evaluates standard statistical summaries. The
calc function however can apply user defined functions over or across raster
layers.
ndvi_std <- sqrt(diag(ndvi_cov))
ndvi_stdz <- calc(ndvi,
function(x) (x - ndvi_mean) / ndvi_std,
filename = file.path(out, 'ndvi_stdz.grd'),
overwrite = TRUE)
Standardizing the data removes the large seasonal swing, but not the correlation between “variables”, i.e. between pixels in different time slices. Only the correlation matters for PCA.
> animate(ndvi_stdz, pause = 0.5, n = 1)
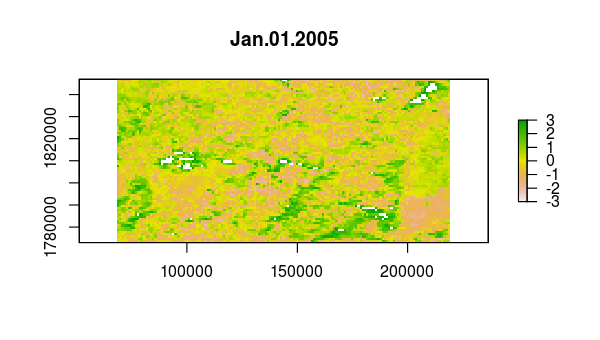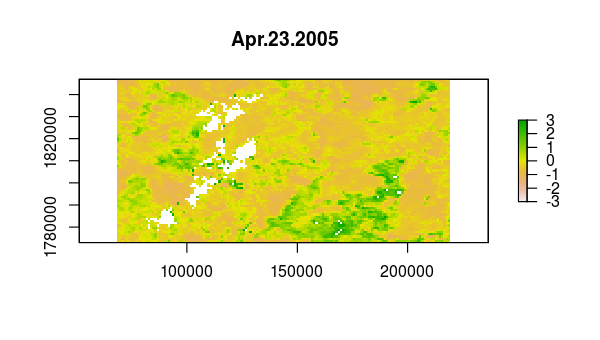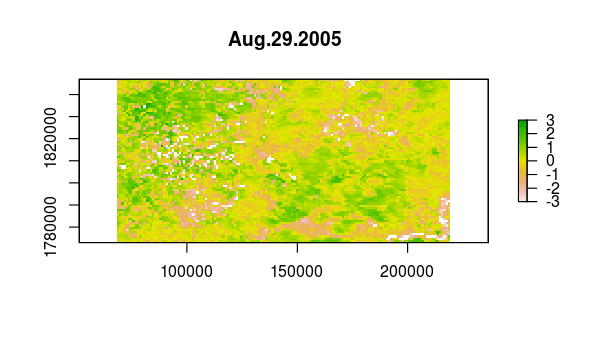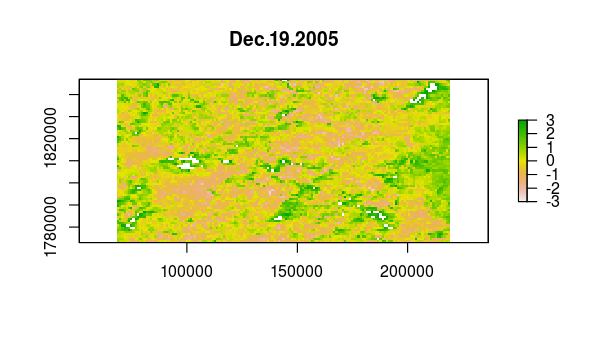
Now, the principal component calculation proceeds using the NDVI correlations, which is just a 23 by 23 matrix of pairwise correlations between the 23 time slices. The plot method of the output shows the variance among pixels, not at each time slice, but on each principal component.
pca <- princomp(covmat = ndvi_cor)
plot(pca)
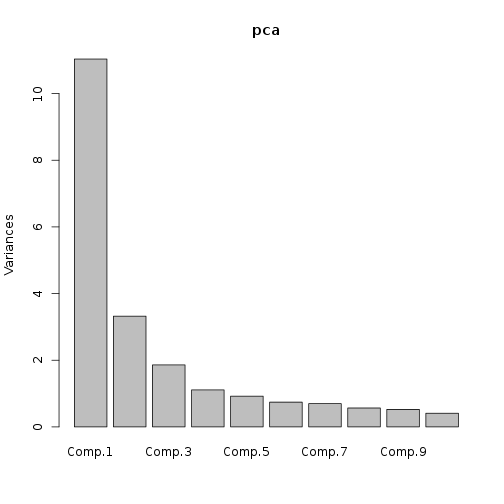
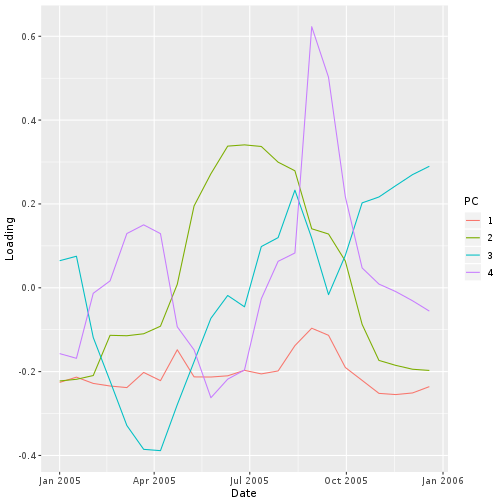
Principal component “loadings” correspond to the weight each time slice contributes to each component.
npc <- 4
loading <- data.frame(
Date = rep(dates, npc),
PC = factor(
rep(1:npc, each = length(dates))
),
Loading = c(pca$loadings[, 1:npc])
)
The first principal component is a more-or-less equally weighted combination of all time slices, like an average.
ggplot(loading,
aes(x = Date, y = Loading,
col = PC)) +
geom_line()
The principal component scores are projections of the NDVI values at each time
point onto the components. Memory limitation may foil a straightforward attempt at this calculation, but the raster
package predict wrapper carries the princomp predict method through to
the time series for each pixel.
pca$center <- pca$scale * 0
ndvi_scores <- predict(
ndvi_stdz, pca,
index = 1:npc,
filename = file.path(out, 'ndvi_scores.grd'),
overwrite = TRUE)
plot(ndvi_scores)
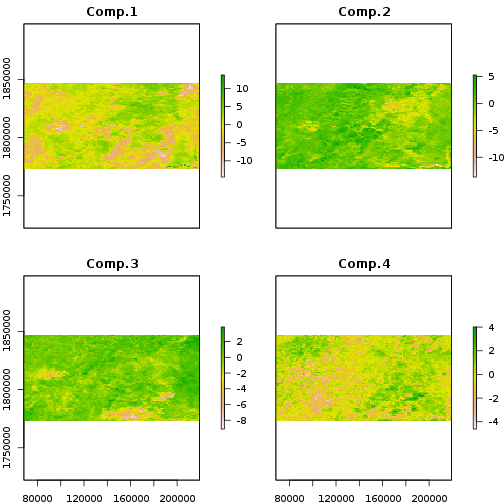
A complication in here is that the pca object does not know how the original
data were centered, because we didn’t give it the original data. The predict
function will behave as if we performed PCA on ndvi_stdz[] if we set the
centering vector to zeros.
The first several principal components account for most of the variance in the data, so approximate the NDVI time series by “un-projecting” the scores.
Mathematically, the calculation for this approximation at each time slice, , is a linear combination of each score “map”, , with time-varying loadings, .
The flexible overlay function allows you to pass a custom function for
pixel-wise calculations on one or more of the main raster objects.
ndvi_dev <- overlay(
ndvi_stdz, ndvi_scores,
fun = function(x, y) {
x - y %*% t(pca$loadings[, 1:npc])
},
filename = file.path(out, 'ndvi_dev.grd'),
overwrite = TRUE)
names(ndvi_dev) <- names(ndvi)
Verify that the deviations just calculated are never very large, then try the same approximation using even fewer principal components.
> animate(ndvi_dev, pause = 0.5, n = 1)
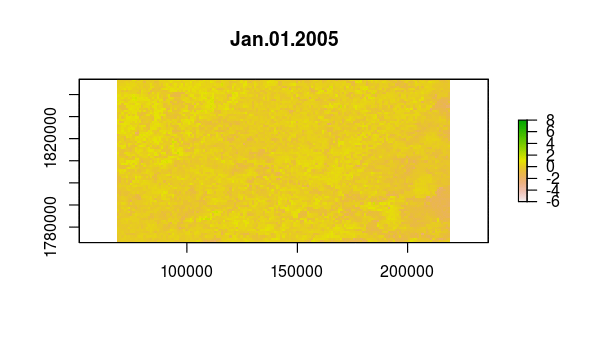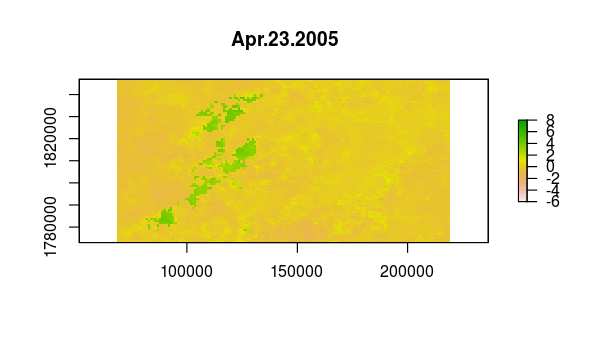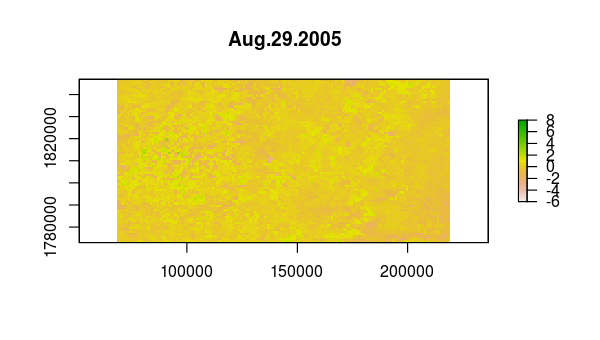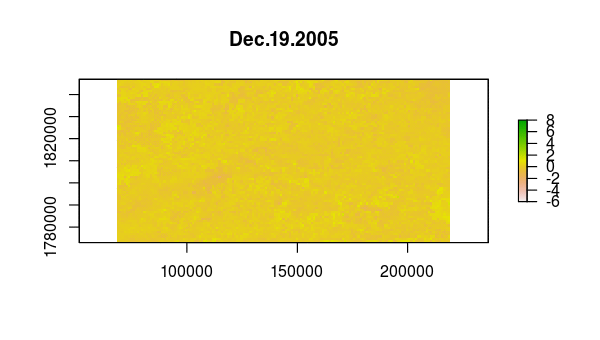
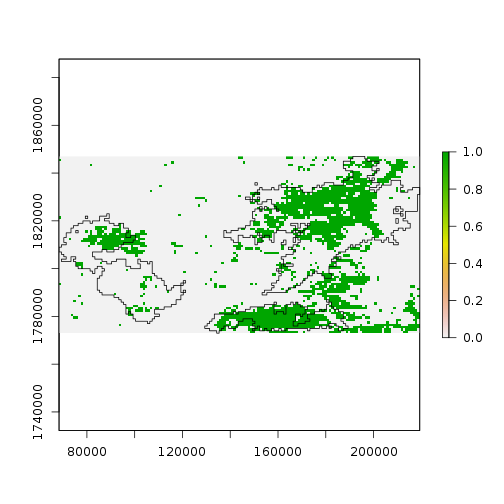
Based on the time variation in the loadings for principal components 2 and 3, we might guess that they correspond to one longer-term and one shorter-term departure from the seasonal NDVI variation within this extent.
plot(
ndvi_scores[[2]] < -2 |
ndvi_scores[[3]] < -2)
plot(st_geometry(scar), add = TRUE)
Summary
The RasterBrick objects created by the brick function, or sometimes as the
result of calculations on a RasterStack, are built for many layers of large
rasters. Key principals for efficient analysis on raster time series include:
- consolidate
RasterStackstoRasterBricksfor faster computation - provide
filenamearguments to keep objects on disk - use
layerStatsto get a covariance matrix for PCA - use
calcandoverlayto call custom functions
If you need to catch-up before a section of code will work, just squish it's 🍅 to copy code above it into your clipboard. Then paste into your interpreter's console, run, and you'll be ready to start in on that section. Code copied by both 🍅 and 📋 will also appear below, where you can edit first, and then copy, paste, and run again.
# Nothing here yet!