Online Data with R
Note: This lesson is in beta status! It may have issues that have not been addressed.
Handouts for this lesson need to be saved on your computer. Download and unzip this material into the directory (a.k.a. folder) where you plan to work.
Lesson Objectives
- Distinguish three ways of acquiring online data
- Break down how web services use HTTP
- Learn R tools for data acquisition
Specific Achievements
- Programatically acquire data embedded in a web page
- Request data through a REST API
- Use the tidycensus package to acquire data
- Use SQLite for caching
Why script data acquisition?
- Too time intensive to acquire manually
- Integrate updated or new data
- Reproducibility
- There’s an API between you and the data
Acquiring Online Data
Data is available on the web in many different forms. How difficult is it to acquire that data to run analyses? It depends which of three approaches the data source requires:
- Web scraping
- Web service (API)
- Specialized package (API wrapper)
Web Scraping 🙁
A web browser reads HTML and JavaScript and displays a human readable page. In contrast, a web scraper is a program (a “bot”) that reads HTML and JavaScript and stores the data.
Web Service (API) 😉
API stands for Application Programming Interface (API, as opposed to GUI) that is compatible with passing data around the internet using HTTP (Hyper-text Transfer Protocol). This is not the fastest protocol for moving large datasets, but it is universal (it underpins web browsers, after all).
Specialized Package 😂
Major data providers can justify writing a “wrapper” package for their API, specific to your language of choice (e.g. Python or R), that facilitates accessing the data they provide through a web service. Sadly, not all do so.
Web Scraping
That “http” at the beginning of the URL for a possible data source is a protocol—an understanding between a client and a server about how to communicate. The client could either be a web browser such as Chrome or Firefox, or your web scraping program written in R, as long as it uses the correct protocol. After all, servers exist to serve.
The following example uses the httr and rvest packages to issue a HTTP request and handle the response.
The page we are scraping, http://research.jisao.washington.edu/pdo/PDO.latest, deals with the Pacific Decadal Oscillation (PDO), a periodic switching between warm and cool water temperatures in the northern Pacific Ocean. Specifically, it contains monthly values from 1900-2018 indicating how far above or below normal the sea surface temperature across the northern Pacific Ocean was during that month.
library(httr)
response <- GET('http://research.jisao.washington.edu/pdo/PDO.latest')
response
Response [http://research.jisao.washington.edu/pdo/PDO.latest]
Date: 2021-12-01 14:50
Status: 200
Content-Type: <unknown>
Size: 12.3 kB
<BINARY BODY>
The GET() function from httr can be used with a single argument,
a text string with the URL of the page you are scraping.
The response is binary (0s and 1s). The rvest package translates
the raw content into an HTML document, just like a browser does. We use the
read_html function to do this.
library(rvest)
pdo_doc <- read_html(response)
pdo_doc
{html_document}
<html>
[1] <body><p>PDO INDEX\n\nIf the columns of the table appear without formatti ...
The HTML document returned by read_html is no longer 0s and 1s, it now
contains readable text. However it is stored as a single long character string.
We need to do some additional processing to make it useful.
If you look at the HTML document, you can see that all the data is inside an
element called "p". We use the html_node function to extract the
single "p" element from the HTML document, then the html_text function
to extract the text from that element.
pdo_node <- html_node(pdo_doc, "p")
pdo_text <- html_text(pdo_node)
The first argument of html_node is the HTML document, and the second
argument is the name of the element we want to extract. html_text
takes the extracted element as input.
Now we have a long text string containing all the data. We can use text mining tools
like regular expressions to pull out data. If we want the twelve monthly
values for the year 2017, we can use the stringr package to get
all the text between the strings “2017” and “2018” with str_match.
library(stringr)
pdo_text_2017 <- str_match(pdo_text, "(?<=2017).*.(?=\\n2018)")
Then extract all the numeric values in the substring with str_extract_all.
str_extract_all(pdo_text_2017[1], "[0-9-.]+")
[[1]]
[1] "0.77" "0.70" "0.74" "1.12" "0.88" "0.79" "0.10" "0.09" "0.32" "0.05"
[11] "0.15" "0.50"
You can learn more about how to use regular expressions to extract information from text strings in SESYNC’s text mining lesson.
Manual web scraping is hard!
Pages designed for humans are increasingly harder to parse programmatically.
- Servers provide different responses based on client “metadata”
- JavaScript often needs to be executed by the client
- The HTML
<table>is drifting into obscurity (mostly for the better)
HTML Tables
Sites with easily accessible HTML tables nowadays may be specifically intended to be parsed programmatically, rather than browsed by a human reader. The US Census provides some documentation for their data services in a massive table:
html_table() converts the HTML table into an R
data frame. Set fill = TRUE so that inconsistent numbers
of columns in each row are filled in.
census_vars_doc <- read_html('https://api.census.gov/data/2017/acs/acs5/variables.html')
table_raw <- html_node(census_vars_doc, 'table')
# This line takes a few moments to run.
census_vars <- html_table(table_raw, fill = TRUE)
> head(census_vars)
# A tibble: 6 × 9
Name Label Concept Required Attributes Limit `Predicate Type` Group ``
<chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 25111… 25111… "25111 v… 25111 v… "25111 va… 2511… 25111 variables 2511… 2511…
2 AIANHH Geogr… "" not req… "" 0 (not a predicat… N/A <NA>
3 AIHHTL Geogr… "" not req… "" 0 (not a predicat… N/A <NA>
4 AIRES Geogr… "" not req… "" 0 (not a predicat… N/A <NA>
5 ANRC Geogr… "" not req… "" 0 (not a predicat… N/A <NA>
6 B0000… Estim… "UNWEIGH… not req… "B00001_0… 0 int B000… <NA>
We can use our tidy data tools to search this unwieldy documentation for variables of interest.
The call to set_tidy_names() is necessary because the table
extraction results in some columns with undefined names—a
common occurrence when parsing Web content. Next, we use select()
to select only the Name and Label columns, and filter()
to select only the rows where the Label column contains the
substring "Median household income". The grepl() function
allows us to filter by a regular expression.
library(tidyverse)
census_vars %>%
set_tidy_names() %>%
select(Name, Label) %>%
filter(grepl('Median household income', Label))
# A tibble: 21 × 2
Name Label
<chr> <chr>
1 B19013_001E Estimate!!Median household income in the past 12 months (in 201…
2 B19013A_001E Estimate!!Median household income in the past 12 months (in 201…
3 B19013B_001E Estimate!!Median household income in the past 12 months (in 201…
4 B19013C_001E Estimate!!Median household income in the past 12 months (in 201…
5 B19013D_001E Estimate!!Median household income in the past 12 months (in 201…
6 B19013E_001E Estimate!!Median household income in the past 12 months (in 201…
7 B19013F_001E Estimate!!Median household income in the past 12 months (in 201…
8 B19013G_001E Estimate!!Median household income in the past 12 months (in 201…
9 B19013H_001E Estimate!!Median household income in the past 12 months (in 201…
10 B19013I_001E Estimate!!Median household income in the past 12 months (in 201…
# … with 11 more rows
Web Services
The US Census Bureau provides access to its vast stores of demographic data over the Web via their API at https://api.census.gov.
The I in GUI is for interface—it’s the same in API, where buttons and drop-down menus are replaced by functions and object attributes.
Instead of interfacing with a user, this kind of interface is suitable for use in programming another software application. In the case of the Census, the main component of the application is some relational database management system. There are several GUIs designed for humans to query the Census database; the Census API is meant for communication between your program (i.e. script) and their application.
You’ll often see the acronym “REST API.” In this context, REST stands for Representational state transfer. This refers to a set of standards that help ensure that the Web service works well with any computer system it may interact with.
The following code acquires data from the US Census Bureau’s American Community Survey (ACS). The ACS is a yearly survey that provides detailed population and housing information at fine geographic scale across the United States. ACS5 refers to a five-year average of the annual surveys.
Look carefully at this URL.
The URL is a query to the US Census API. The parameters after the ?
request the variable NAME for all counties in state 24 (Maryland).
In a web service, the already universal system for transferring data over the internet, known as HTTP, is half of the interface. All you really need is documentation for how to construct the URL in a standards-compliant way that the service will recognize.
| Section | Description |
|---|---|
https:// |
scheme |
api.census.gov |
authority, or simply domain if there’s no user authentication |
/data/2018/acs/acs5 |
path to a resource within a hierarchy |
? |
beginning of the query component of a URL |
get=NAME |
first query parameter |
& |
query parameter separator |
for=county:* |
second query parameter |
& |
query parameter separator |
in=state:24 |
third query parameter |
# |
beginning of the fragment component of a URL |
irrelephant |
a document section, it isn’t even sent to the server |
To construct the URL in R and send the query to the API, use GET() from
httr.
The first argument to GET() is the base URL, and the
query argument is a named list that passes
the parameters of the query to the API. All the elements
of the list should be character strings.
path <- 'https://api.census.gov/data/2018/acs/acs5'
query_params <- list('get' = 'NAME,B19013_001E',
'for' = 'county:*',
'in' = 'state:24')
response <- GET(path, query = query_params)
response
Response [https://api.census.gov/data/2018/acs/acs5?get=NAME%2CB19013_001E&for=county%3A%2A&in=state%3A24]
Date: 2021-12-01 14:50
Status: 200
Content-Type: application/json;charset=utf-8
Size: 1.25 kB
[["NAME","B19013_001E","state","county"],
["Howard County, Maryland","117730","24","027"],
["Prince George's County, Maryland","81969","24","033"],
["Anne Arundel County, Maryland","97810","24","003"],
["Baltimore County, Maryland","74127","24","005"],
["Frederick County, Maryland","91999","24","021"],
["Calvert County, Maryland","104301","24","009"],
["Garrett County, Maryland","49619","24","023"],
["Kent County, Maryland","56009","24","029"],
["Montgomery County, Maryland","106287","24","031"],
...
Response Header
The response from the API is a bunch of 0s and 1s, but part of the HTTP protocol is to include a “header” with information about how to decode the body of the response.
The output of GET() has already decoded the body of the response
from binary format.
Most REST APIs return as the “content” either:
- Javascript Object Notation (JSON)
- a UTF-8 encoded string of key-value pairs, where values may be lists
- e.g.
{'a':24, 'b': ['x', 'y', 'z']}
- eXtensible Markup Language (XML)
- a nested
<tag></tag>hierarchy serving the same purpose
- a nested
The header from Census says the content type is JSON.
response$headers['content-type']
$`content-type`
[1] "application/json;charset=utf-8"
Response Content
First, use httr::content() to retrieve
the JSON content of the response. Use as = 'text' to
get the content as a character vector. Then use
jsonlite::fromJSON() to convert to a matrix.
library(jsonlite)
county_income <- response %>%
content(as = 'text') %>%
fromJSON()
> county_income
[,1] [,2] [,3] [,4]
[1,] "NAME" "B19013_001E" "state" "county"
[2,] "Howard County, Maryland" "117730" "24" "027"
[3,] "Prince George's County, Maryland" "81969" "24" "033"
[4,] "Anne Arundel County, Maryland" "97810" "24" "003"
[5,] "Baltimore County, Maryland" "74127" "24" "005"
[6,] "Frederick County, Maryland" "91999" "24" "021"
[7,] "Calvert County, Maryland" "104301" "24" "009"
[8,] "Garrett County, Maryland" "49619" "24" "023"
[9,] "Kent County, Maryland" "56009" "24" "029"
[10,] "Montgomery County, Maryland" "106287" "24" "031"
[11,] "Carroll County, Maryland" "93363" "24" "013"
[12,] "Queen Anne's County, Maryland" "92167" "24" "035"
[13,] "St. Mary's County, Maryland" "90438" "24" "037"
[14,] "Charles County, Maryland" "95924" "24" "017"
[15,] "Dorchester County, Maryland" "52145" "24" "019"
[16,] "Washington County, Maryland" "59719" "24" "043"
[17,] "Wicomico County, Maryland" "56608" "24" "045"
[18,] "Cecil County, Maryland" "72845" "24" "015"
[19,] "Caroline County, Maryland" "54956" "24" "011"
[20,] "Worcester County, Maryland" "61145" "24" "047"
[21,] "Talbot County, Maryland" "67204" "24" "041"
[22,] "Baltimore city, Maryland" "48840" "24" "510"
[23,] "Somerset County, Maryland" "42165" "24" "039"
[24,] "Harford County, Maryland" "85942" "24" "025"
[25,] "Allegany County, Maryland" "44065" "24" "001"
Notice that the matrix created by fromJSON() does not recognize that the first
row is a header, resulting in all columns being classified as
character. This is a typical situation when parsing Web content, and would require
additional data wrangling to fix.
API Keys & Limits
Most servers request good behavior, others enforce it.
- Size of single query
- Rate of queries (calls per second, or per day)
- User credentials specified by an API key
From the Census FAQ What Are the Query Limits?:
You can include up to 50 variables in a single API query and can make up to 500 queries per IP address per day… Please keep in mind that all queries from a business or organization having multiple employees might employ a proxy service or firewall. This will make all of the users of that business or organization appear to have the same IP address.
Specialized Packages
The third tier of access to online data is much preferred, if it exists: a dedicated package in your programming language’s repository, CRAN or PyPI.
- Additional guidance on query parameters
- Returns data in native formats
- Handles all “encoding” problems
The tidycensus package, developed by Kyle Walker, streamlines access to the API and is integrated with tidyverse packages.
To repeat the exercise below at home, request an API key at
https://api.census.gov/data/key_signup.html, and store it in a file named census_api_key.R
in your working directory. The file should contain the line
Sys.setenv(CENSUS_API_KEY = 'your many digit key'). This creates a hidden
system variable containing the key. This is good practice—it is much safer than
pasting the API key directly into your code or saving it as a variable in the global environment.
library(tidycensus)
> source('census_api_key.R')
Compared to using the API directly via the httr package:
Pros
- More concise code, quicker development
- Package documentation (if present) is usually more user-friendly than API documentaion.
- May allow seamless update if API changes
Cons
- No guarantee of updates
- Possibly limited in scope
Query the Census ACS5 survey for the variable B19013_001E (median annual household income,
in dollars) and each entity’s NAME.
variables <- c('NAME', 'B19013_001E')
Get the variables NAME and B19013_001E (median household income)
from all counties in Maryland. tidycensus
converts the JSON string into a data frame. (No need to check headers.)
This code uses the get_acs function, which is the main function in
tidycensus for interacting with the American Community Survey
API. The arguments are fairly self-explanatory. We can use the text
abbreviation for the state of Maryland (MD); the function automatically
converts this into the numerical FIPS code. The geometry = TRUE argument
means that we want get_acs output to include the county boundaries as a
spatial object, to easily create maps of our data.
county_income <- get_acs(geography = 'county',
variables = variables,
state = 'MD',
year = 2018,
geometry = TRUE)
> county_income
Simple feature collection with 24 features and 5 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: -79.48765 ymin: 37.91172 xmax: -75.04894 ymax: 39.72304
CRS: 4269
# A tibble: 24 × 6
GEOID NAME variable estimate moe geometry
<chr> <chr> <chr> <dbl> <dbl> <MULTIPOLYGON [°]>
1 24001 Allegany … B19013_0… 44065 1148 (((-79.06756 39.47944, -79.06003 3…
2 24003 Anne Arun… B19013_0… 97810 1299 (((-76.84036 39.10314, -76.83678 3…
3 24005 Baltimore… B19013_0… 74127 922 (((-76.3257 39.31397, -76.32452 39…
4 24009 Calvert C… B19013_0… 104301 3548 (((-76.70121 38.71276, -76.69915 3…
5 24011 Caroline … B19013_0… 54956 2419 (((-76.01505 38.72869, -76.01321 3…
6 24013 Carroll C… B19013_0… 93363 1867 (((-77.31151 39.63914, -77.30972 3…
7 24015 Cecil Cou… B19013_0… 72845 2208 (((-76.23326 39.72131, -76.15435 3…
8 24017 Charles C… B19013_0… 95924 2651 (((-77.27382 38.48356, -77.2724 38…
9 24019 Dorcheste… B19013_0… 52145 4243 (((-76.06544 38.1298, -76.06461 38…
10 24021 Frederick… B19013_0… 91999 1580 (((-77.67716 39.32453, -77.67637 3…
# … with 14 more rows
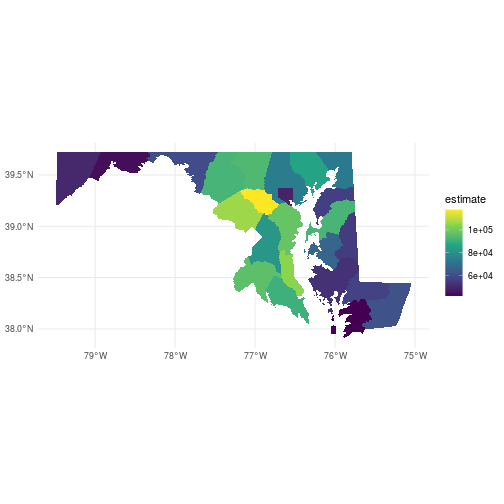
We can use dplyr to manipulate the output, and ggplot2 to visualize the data.
Because we set geometry = TRUE, tidycensus even includes spatial information in its
output that we can use to create maps!
This code uses the spatial data frame output from get_acs to plot the counties of Maryland with
fill color corresponding to the median household income of the counties, with some additional
graphical options.
ggplot(county_income) +
geom_sf(aes(fill = estimate), color = NA) +
coord_sf() +
theme_minimal() +
scale_fill_viridis_c()
For a more in-depth tutorial on R’s geospatial data types, check out SESYNC’s lesson on geospatial packages in R.
Paging & Stashing
A common strategy that web service providers take to balance their load is to limit the number of records a single API request can return. The user ends up having to flip through “pages” with the API, handling the response content at each iteration. Options for stashing data are:
- Store it all in memory, write to file at the end.
- Append each response to a file, writing frequently.
- Offload these decisions to database management software.
The data.gov API provides a case in point. Data.gov is a service provided by the U.S. federal government to make data available from across many government agencies. It hosts a catalog of raw data and of many other APIs from across government. Among the APIs catalogued by data.gov is the FoodData Central API. The U.S. Department of Agriculture maintains a data system of nutrition information for thousands of foods. We might be interested in the relative nutrient content of different fruits.
To repeat the exercise below at home, request an API key at
https://api.data.gov/signup/, and store it in a file named datagov_api_key.R
in your working directory. The file should contain the line
Sys.setenv(DATAGOV_KEY = 'your many digit key').
Load the DATAGOV_KEY variable as a system variable by importing it from the file you saved it in.
> source('datagov_api_key.R')
Run an API query for all foods with "fruit" in their name and parse the content of the response.
Just like we did previously in this lesson, we create a named list
of query parameters, including the API key and the
search string, and pass them to GET(). We use the pipe operator %>%
to pipe the output of GET() to content(). We use the as = 'parsed'
argument to convert the JSON content to a nested list.
api <- 'https://api.nal.usda.gov/fdc/v1/'
path <- 'foods/search'
query_params <- list('api_key' = Sys.getenv('DATAGOV_KEY'),
'query' = 'fruit')
doc <- GET(paste0(api, path), query = query_params) %>%
content(as = 'parsed')
Extract data from the returned JSON object, which gets mapped to an
R list called doc.
First inspect the names of the list elements.
> names(doc)
[1] "totalHits" "currentPage" "totalPages"
[4] "pageList" "foodSearchCriteria" "foods"
[7] "aggregations"
We can print the value of doc$totalHits to see
how many foods matched our search term, "fruit".
> doc$totalHits
[1] 38999
The claimed number of results is much larger than the length
of the foods array contained in this response. The query returned only the
first page, with 50 items.
> length(doc$foods)
[1] 50
Continue to inspect the returned object. Extract one element from the list
of foods and view its description.
> fruit <- doc$foods[[1]]
> fruit$description
[1] "Fruit leather and fruit snacks candy"
The map_dfr function from the purrr package extracts the name and
value of all the nutrients in the foodNutrients list within the first search
result, and creates a data frame.
nutrients <- map_dfr(fruit$foodNutrients,
~ data.frame(name = .$nutrientName,
value = .$value))
> head(nutrients, 10)
name value
1 Protein 0.55
2 Total lipid (fat) 2.84
3 Carbohydrate, by difference 84.30
4 Energy 365.00
5 Alcohol, ethyl 0.00
6 Water 11.20
7 Caffeine 0.00
8 Theobromine 0.00
9 Sugars, total including NLEA 53.40
10 Fiber, total dietary 0.00
The map_dfr function applies a function to each element in foodNutrients, which
returns a single-row data frame with two columns. Then, it combines all 50 of
the single-row data frames to create a 50-row data frame and returns it.
The DBI and RSQLite packages together allow R to connect to a
database-in-a-file. If the fruits.sqlite file does not exist
in your working directory already when you try to connect,
dbConnect() will create it.
library(DBI)
library(RSQLite)
fruit_db <- dbConnect(SQLite(), 'fruits.sqlite')
Add a new pageSize parameter by appending a named element
to the existing query_params list, to request 100 documents per page.
query_params$pageSize <- 100
We will send 10 queries to the API to get 1000 total records.
In each request (each iteration through the loop),
advance the query parameter pageNumber by one.
The query will retrieve 100 records, starting with pageNumber * pageSize.
We use some tidyr and dplyr manipulations to
extract the ID number, name, and the amount of sugar from each
of the foods in the page of results returned by the query. The series of
unnest_longer() and unnest_wider() functions turns the nested list into
a data frame by successively converting lists into columns in the data frame.
This manipulation is necessary because R does not easily handle the
nested list structures that APIs return. If we were using
a specialized API R package, typically it would handle this data wrangling
for us. After converting the list to a data frame, we use filter to retain
only the rows where the nutrientName contains the substring 'Sugars, total'
and then select the three columns we want to keep: the numerical ID of
the food, its full name, and its sugar content. Finally the 100-row data
frame is assigned to the object values.
Each time through the loop, insert the next 100 fruits
(the three-column data frame values)
in bulk to the database with dbWriteTable().
for (i in 1:10) {
# Advance page and query
query_params$pageNumber <- i
response <- GET(paste0(api, path), query = query_params)
page <- content(response, as = 'parsed')
# Convert nested list to data frame
values <- tibble(food = page$foods) %>%
unnest_wider(food) %>%
unnest_longer(foodNutrients) %>%
unnest_wider(foodNutrients) %>%
filter(grepl('Sugars, total', nutrientName)) %>%
select(fdcId, description, value) %>%
setNames(c('foodID', 'name', 'sugar'))
# Stash in database
dbWriteTable(fruit_db, name = 'Food', value = values, append = TRUE)
}
The first time through the loop, the Food table does not exist in
the database yet, so dbWriteTable() creates it. In subsequent iterations,
the append = TRUE argument tells dbWriteTable() to add new rows to
the existing database.
View the records in the database by reading
everything we have so far into a data frame
with dbReadTable().
fruit_sugar_content <- dbReadTable(fruit_db, name = 'Food')
> head(fruit_sugar_content, 10)
foodID name sugar
1 1104047 Fruit leather and fruit snacks candy 53.40
2 1097696 Fruit smoothie, with whole fruit and dairy 8.23
3 1102761 Fruit smoothie, with whole fruit, no dairy 8.20
4 1103961 Topping, fruit 27.40
5 1102738 Soup, fruit 5.87
6 1100841 Biscuit with fruit 10.00
7 1100884 Bread, fruit 24.80
8 167781 Candied fruit 80.70
9 1100904 Cheesecake with fruit 15.30
10 1100710 Croissant, fruit 14.10
Don’t forget to disconnect from your database!
dbDisconnect(fruit_db)
Takeaway
-
Web scraping is hard and unreliable, but sometimes there is no other option.
-
Web services are the most common resource.
-
Use a package specific to your API if one is available.
Web services do not always have great documentation—what parameters are acceptable or necessary may not be clear. Some may even be poorly documented on purpose if the API wasn’t designed for public use! Even if you plan to acquire data using the “raw” web service, try a search for a relevant package on CRAN. The package documentation could help.
For more resources, and to discover more APIs, visit:
- DataOne
A network of data repositories making data more accessible and usable.- Data Discovery: Portal to search DataOne
- metajam: R package to help you access data on DataOne
-
SODA Developers
An open data API to access open data resources from many organizations and governments. - Public APIs
A repository listing free APIs for use in your research and projects.
A final note on U.S. Census packages: In this lesson, we use Kyle Walker’s tidycensus package, but you might also want to take a look at Hannah Recht’s censusapi or Ezra Glenn’s acs. All three packages take slightly different approaches to obtaining data from the U.S. Census API.
Exercises
Exercise 1
Create a data frame with the population of all countries in the world by scraping
the Wikipedia list of countries by population.
Hint: First call the function read_html(), then call html_node()
on the output of read_html() with the argument xpath='//*[@id="mw-content-text"]/div/table[1]'
to extract the table element from the HTML content, then call a third function to
convert the HTML table to a data frame.
Exercise 2
Identify the name of the census variable in the table of ACS variables whose “Concept” column includes “COUNT OF THE POPULATION”. Next, use the Census API to collect the data for this variable, for every county in Maryland (FIPS code 24) into a data frame. Optional: Create a map or figure to visualize the data.
Exercise 3
Request an API key for data.gov, which will enable you to access the FoodData Central API. Use the API to collect 3 “pages” of food results matching a search term of your choice. Save the names of the foods and a nutrient value of your choice into a new SQLite file.
Solutions
Solution 1
> library(rvest)
> url <- 'https://en.wikipedia.org/wiki/List_of_countries_and_dependencies_by_population'
> doc <- read_html(url)
> table_node <- html_node(doc, xpath='//*[@id="mw-content-text"]/div/table[1]')
> pop_table <- html_table(table_node)
Solution 2
> library(tidyverse)
> library(tidycensus)
> source('census_api_key.R')
>
> # Using the previously created census_vars table, find the variable ID for population count.
> census_vars <- set_tidy_names(census_vars)
> population_vars <- census_vars %>%
+ filter(grepl('COUNT OF THE POPULATION', Concept))
> pop_var_id <- population_vars$Name[1]
>
> # Use tidycensus to query the API.
> county_pop <- get_acs(geography = 'county',
+ variables = pop_var_id,
+ state = 'MD',
+ year = 2018,
+ geometry = TRUE)
>
> # Map of counties by population
> ggplot(county_pop) +
+ geom_sf(aes(fill = estimate), color = NA) +
+ coord_sf() +
+ theme_minimal() +
+ scale_fill_viridis_c()
Solution 3
Here is a possible solution getting the protein content from different kinds of cheese.
> library(httr)
> library(DBI)
> library(RSQLite)
>
> source('datagov_api_key.R')
>
> api <- 'https://api.nal.usda.gov/fdc/v1/'
> path <- 'foods/search'
>
> query_params <- list('api_key' = Sys.getenv('DATAGOV_KEY'),
+ 'query' = 'cheese',
+ 'pageSize' = 100)
>
> # Create a new database
> cheese_db <- dbConnect(SQLite(), 'cheese.sqlite')
>
> for (i in 1:3) {
+ # Advance page and query
+ query_params$pageNumber <- i
+ response <- GET(paste0(api, path), query = query_params)
+ page <- content(response, as = 'parsed')
+
+ # Convert nested list to data frame
+ values <- tibble(food = page$foods) %>%
+ unnest_wider(food) %>%
+ unnest_longer(foodNutrients) %>%
+ unnest_wider(foodNutrients) %>%
+ filter(grepl('Protein', nutrientName)) %>%
+ select(fdcId, description, value) %>%
+ setNames(c('foodID', 'name', 'protein'))
+
+ # Stash in database
+ dbWriteTable(cheese_db, name = 'Food', value = values, append = TRUE)
+
+ }
>
> dbDisconnect(cheese_db)
If you need to catch-up before a section of code will work, just squish it's 🍅 to copy code above it into your clipboard. Then paste into your interpreter's console, run, and you'll be ready to start in on that section. Code copied by both 🍅 and 📋 will also appear below, where you can edit first, and then copy, paste, and run again.
# Nothing here yet!